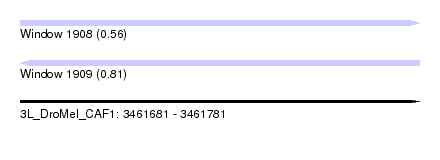
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,461,681 – 3,461,781 |
| Length | 100 |
| Max. P | 0.808986 |

| Location | 3,461,681 – 3,461,781 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -14.05 |
| Consensus MFE | -11.96 |
| Energy contribution | -12.03 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3461681 100 + 23771897 UGGCCAGGGAAAAUUCAAUCAACGUUUUGUUUUUAAUCGAUUCCUAUUGCAAUUCCCAGCUUUUCUCUCCUUUUCUUGGCUUUUCCCUUAGCUUAUUUCU .(((.((((((((..(((.....(..(((........)))..).....((........))...............)))..))))))))..)))....... ( -14.50) >DroSec_CAF1 9010 99 + 1 UGGCCAGGGAAAAUUCAAUCAACGUUUUGUUUUUAAUCGAUUCCUAUUGCAAUCCCCAGUUUUCCCC-CCCUUUCUUGGCUUUUCCCUUAGCUUACUUCU .(((((((((((((((((........))).........((((.(....).))))...))))))))).-........)))))................... ( -14.40) >DroSim_CAF1 9031 100 + 1 UGGCCAGGGAAAAUUCAAUCAACGUUUUGUUUUUAAUCGAUUCCUAUUGCAAUCCCCAGCUUUCCCCUCCCUUUCUUGGCUUUUCCCUUAGCUUACUUCU .(((.((((((((..(((.....(..(((........)))..).....((........))...............)))..))))))))..)))....... ( -14.50) >DroYak_CAF1 9259 100 + 1 UGGCCAGGGAAAAUUCAAUCAACGUUUUGUUUUUAAUCGAUUCCUAUUGCAAUCCCCUCUUUACCCCAGCUUUUCUUCGCUUUUCCCUUAGCUUUUUUCU .(((.((((((((.........................((((.(....).))))..............((........))))))))))..)))....... ( -12.80) >consensus UGGCCAGGGAAAAUUCAAUCAACGUUUUGUUUUUAAUCGAUUCCUAUUGCAAUCCCCAGCUUUCCCCUCCCUUUCUUGGCUUUUCCCUUAGCUUACUUCU .(((.((((((((.((((..(((.....))).......((((.(....).)))).....................)))).))))))))..)))....... (-11.96 = -12.03 + 0.06)

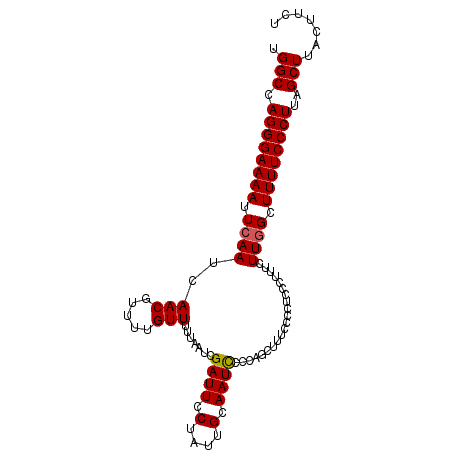
| Location | 3,461,681 – 3,461,781 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -20.40 |
| Consensus MFE | -16.09 |
| Energy contribution | -16.46 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3461681 100 - 23771897 AGAAAUAAGCUAAGGGAAAAGCCAAGAAAAGGAGAGAAAAGCUGGGAAUUGCAAUAGGAAUCGAUUAAAAACAAAACGUUGAUUGAAUUUUCCCUGGCCA ........((((.(((((((....................((........))........((((((((..........)))))))).))))))))))).. ( -19.00) >DroSec_CAF1 9010 99 - 1 AGAAGUAAGCUAAGGGAAAAGCCAAGAAAGGG-GGGGAAAACUGGGGAUUGCAAUAGGAAUCGAUUAAAAACAAAACGUUGAUUGAAUUUUCCCUGGCCA ........((((.(((((((..(((..((.(.-((......))...((((.(....).))))..............).))..)))..))))))))))).. ( -21.20) >DroSim_CAF1 9031 100 - 1 AGAAGUAAGCUAAGGGAAAAGCCAAGAAAGGGAGGGGAAAGCUGGGGAUUGCAAUAGGAAUCGAUUAAAAACAAAACGUUGAUUGAAUUUUCCCUGGCCA ........(((........))).......((..(((((((((........))........((((((((..........))))))))..)))))))..)). ( -20.20) >DroYak_CAF1 9259 100 - 1 AGAAAAAAGCUAAGGGAAAAGCGAAGAAAAGCUGGGGUAAAGAGGGGAUUGCAAUAGGAAUCGAUUAAAAACAAAACGUUGAUUGAAUUUUCCCUGGCCA ........((((.(((((((...........(((..((((........))))..)))...((((((((..........)))))))).))))))))))).. ( -21.20) >consensus AGAAAUAAGCUAAGGGAAAAGCCAAGAAAAGGAGGGGAAAACUGGGGAUUGCAAUAGGAAUCGAUUAAAAACAAAACGUUGAUUGAAUUUUCCCUGGCCA ........((((.(((((((((.......((..........)).......))........((((((((..........)))))))).))))))))))).. (-16.09 = -16.46 + 0.37)

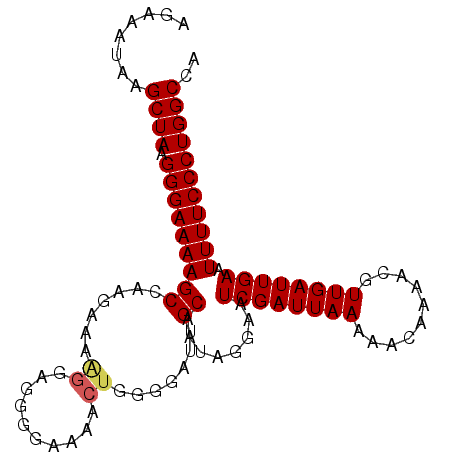
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:18 2006