
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,432,642 – 3,432,736 |
| Length | 94 |
| Max. P | 0.767606 |

| Location | 3,432,642 – 3,432,736 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.36 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -18.83 |
| Energy contribution | -18.67 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
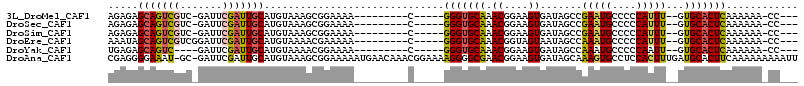
>3L_DroMel_CAF1 3432642 94 + 23771897 AGAGAGCAGUCGUC-GAUUCGAUUGCAUGUAAAGCGGAAAA---------C-----GGGUGCAAACGGAAGUGAUAGCCGAAUGCCCCCAUUU--GUGCACUCAAAAAA-CC--- .(((.(((((((..-....))))))).......(((.(((.---------.-----(((.(((..(((.........)))..))))))..)))--.))).)))......-..--- ( -29.90) >DroSec_CAF1 47983 94 + 1 AGAGAGCAGUCGUC-GAUUCGAUUGCAUGUAAAGCGGAAAA---------C-----GGGUGCAAACGGAAGUGAUAGCCGAAUGCCCCCAUUU--GUGCACUCAAAAAA-CC--- .(((.(((((((..-....))))))).......(((.(((.---------.-----(((.(((..(((.........)))..))))))..)))--.))).)))......-..--- ( -29.90) >DroSim_CAF1 37141 94 + 1 AGAGAGCAGUCGUC-GAUUCGAUUGCAUGUAAAGCGGAAAA---------C-----GGGUGCAAACGGAAGUGAUAGCCGAAUGCCCCCAUUU--GUGCACUCAAAAAA-CC--- .(((.(((((((..-....))))))).......(((.(((.---------.-----(((.(((..(((.........)))..))))))..)))--.))).)))......-..--- ( -29.90) >DroEre_CAF1 49913 95 + 1 AAAUAGCAGUCGUCGGAUUCGAUUGCAUGUAAAACGAAAAA---------C-----GGGUGCAAACGGUAGUAAUAGCCAAAUGCCCCCAUUU--GUGCACUCAAAAAA-CC--- .....(((((((.......)))))))...............---------.-----(((((((...(.((....)).)((((((....)))))--))))))))......-..--- ( -24.60) >DroYak_CAF1 47208 91 + 1 UGAGAGCAGUC----GAUUCGAUUGCAUGUAAAACGGAAAA---------C-----GGGUGCAAACGGAAGUGAUAGCCAAAUGCCCCCAAUU--GUGCACUCAAAAAA-CC--- ((((.((((((----(...))))))).((((...((.....---------)-----)(((....((....))....)))..............--.)))))))).....-..--- ( -23.00) >DroAna_CAF1 78094 113 + 1 CGAGGGGAAAU-GC-GAUUCGAUUGCAUGUAAAGCGGAAAAAUGAACAAACGGAAAAGGGGCGAACGGAAGUGAUAGCAAAGUGCCUCCACUUUGAUGCACUUCAAAAAAAAAUU ((.......((-((-(((...)))))))......))...............................((((((....(((((((....)))))))...))))))........... ( -22.42) >consensus AGAGAGCAGUCGUC_GAUUCGAUUGCAUGUAAAGCGGAAAA_________C_____GGGUGCAAACGGAAGUGAUAGCCAAAUGCCCCCAUUU__GUGCACUCAAAAAA_CC___ .....(((((((.......)))))))..............................(((((((.((....)).......(((((....)))))...)))))))............ (-18.83 = -18.67 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:07 2006