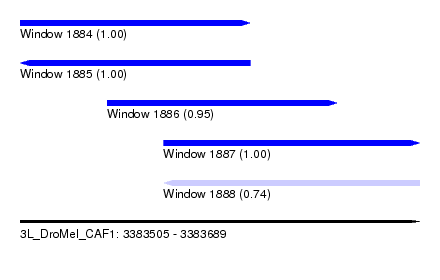
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,383,505 – 3,383,689 |
| Length | 184 |
| Max. P | 0.999986 |

| Location | 3,383,505 – 3,383,611 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.97 |
| Mean single sequence MFE | -37.66 |
| Consensus MFE | -36.08 |
| Energy contribution | -36.48 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.96 |
| SVM decision value | 5.42 |
| SVM RNA-class probability | 0.999986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3383505 106 + 23771897 ACUGUUUGUAAUUCGCGUUGAGCGCGCGAAUUAUGAUUGAAUAAUGAAAUGCCCCAAUGGGCAAU--------------GGGCAAUGGGCAGUCAGUCGCUGCCUUCAAUUAUGCAAAAC ....((((((((((((((.....)))))))))((((((((.........(((((....)))))..--------------.......(((((((.....)))))))))))))))))))).. ( -37.50) >DroSec_CAF1 50969 106 + 1 ACUGUUUGUAAUUCGCGUUGAGCGCGCGAAUUAUGAUUGAAUAAUGAAAUGCCCCAAUGGGCAAU--------------GGGCAAUGGGCAGUCAGUCGCUGCCUUCAAUUAUGCAAAAC ....((((((((((((((.....)))))))))((((((((.........(((((....)))))..--------------.......(((((((.....)))))))))))))))))))).. ( -37.50) >DroSim_CAF1 49129 106 + 1 ACUGUUUGUAAUUCGCGUUGAGCGCGCGAAUUAUGAUUGAAUAAUGAAAUGCCCCAAUGGGCAAU--------------GGGCAAUGGGCAGUCAGUCGCUGCCUUCAAUUAUGCAAAAC ....((((((((((((((.....)))))))))((((((((.........(((((....)))))..--------------.......(((((((.....)))))))))))))))))))).. ( -37.50) >DroEre_CAF1 49932 113 + 1 ACUGCUUGUAAUUCGCGUUGAGCGCGCGAAUUAUGAUUGAAUAAUGAAAUGCCCCAAUGGGC-------AAUGGGUAAUGGGCAAUGGGCAGUCAGUCGCUGCCUUCAAUUAUGCAAAAC ..(((..(((((((((((.....)))))))))))((((((.........(((((....))))-------)................(((((((.....)))))))))))))..))).... ( -37.90) >DroYak_CAF1 49902 120 + 1 ACUGUUUGUAAUUCGCGUUGAGCGCGCGAAUUAUGAUUGAAUAAUGAAAUGCCCCAAUGGGCAAUGGGCAAUGGGUAAUGGGCAAUGGGCAGUCAGUCGCUGCCUUCAAUUAUGCAAAAC ....((((((((((((((.....)))))))))((((((((.........(((((....)))))....((.((.....))..))...(((((((.....)))))))))))))))))))).. ( -37.90) >consensus ACUGUUUGUAAUUCGCGUUGAGCGCGCGAAUUAUGAUUGAAUAAUGAAAUGCCCCAAUGGGCAAU______________GGGCAAUGGGCAGUCAGUCGCUGCCUUCAAUUAUGCAAAAC ....((((((((((((((.....)))))))))((((((((.........(((((....))))).......................(((((((.....)))))))))))))))))))).. (-36.08 = -36.48 + 0.40)


| Location | 3,383,505 – 3,383,611 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.97 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -29.46 |
| Energy contribution | -29.30 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.23 |
| SVM RNA-class probability | 0.999842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3383505 106 - 23771897 GUUUUGCAUAAUUGAAGGCAGCGACUGACUGCCCAUUGCCC--------------AUUGCCCAUUGGGGCAUUUCAUUAUUCAAUCAUAAUUCGCGCGCUCAACGCGAAUUACAAACAGU ((((((.(((((.((((((((.......)))))........--------------..(((((....))))).)))))))).))....(((((((((.......))))))))).))))... ( -30.00) >DroSec_CAF1 50969 106 - 1 GUUUUGCAUAAUUGAAGGCAGCGACUGACUGCCCAUUGCCC--------------AUUGCCCAUUGGGGCAUUUCAUUAUUCAAUCAUAAUUCGCGCGCUCAACGCGAAUUACAAACAGU ((((((.(((((.((((((((.......)))))........--------------..(((((....))))).)))))))).))....(((((((((.......))))))))).))))... ( -30.00) >DroSim_CAF1 49129 106 - 1 GUUUUGCAUAAUUGAAGGCAGCGACUGACUGCCCAUUGCCC--------------AUUGCCCAUUGGGGCAUUUCAUUAUUCAAUCAUAAUUCGCGCGCUCAACGCGAAUUACAAACAGU ((((((.(((((.((((((((.......)))))........--------------..(((((....))))).)))))))).))....(((((((((.......))))))))).))))... ( -30.00) >DroEre_CAF1 49932 113 - 1 GUUUUGCAUAAUUGAAGGCAGCGACUGACUGCCCAUUGCCCAUUACCCAUU-------GCCCAUUGGGGCAUUUCAUUAUUCAAUCAUAAUUCGCGCGCUCAACGCGAAUUACAAGCAGU ...((((...(((((((((((.......))))).................(-------((((....)))))........))))))..(((((((((.......)))))))))...)))). ( -32.60) >DroYak_CAF1 49902 120 - 1 GUUUUGCAUAAUUGAAGGCAGCGACUGACUGCCCAUUGCCCAUUACCCAUUGCCCAUUGCCCAUUGGGGCAUUUCAUUAUUCAAUCAUAAUUCGCGCGCUCAACGCGAAUUACAAACAGU ((((((.(((((.((((((((.......)))))........................(((((....))))).)))))))).))....(((((((((.......))))))))).))))... ( -30.00) >consensus GUUUUGCAUAAUUGAAGGCAGCGACUGACUGCCCAUUGCCC______________AUUGCCCAUUGGGGCAUUUCAUUAUUCAAUCAUAAUUCGCGCGCUCAACGCGAAUUACAAACAGU ((((((.(((((.((((((((.......))))).........................((((....))))..)))))))).))....(((((((((.......))))))))).))))... (-29.46 = -29.30 + -0.16)


| Location | 3,383,545 – 3,383,651 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.32 |
| Mean single sequence MFE | -28.10 |
| Consensus MFE | -26.30 |
| Energy contribution | -26.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3383545 106 + 23771897 AUAAUGAAAUGCCCCAAUGGGCAAU--------------GGGCAAUGGGCAGUCAGUCGCUGCCUUCAAUUAUGCAAAACGGUUACUGCACUUUGCAUUAAUUAACAAACGGGCAAACAA .........(((((....))))).(--------------(.((...(((((((.....)))))))..(((((((((((...((....))..)))))).))))).........))...)). ( -27.60) >DroSec_CAF1 51009 106 + 1 AUAAUGAAAUGCCCCAAUGGGCAAU--------------GGGCAAUGGGCAGUCAGUCGCUGCCUUCAAUUAUGCAAAACGGUUACUGCACUUUGCAUUAAUUAACAAACGGGCAAACAA .........(((((....))))).(--------------(.((...(((((((.....)))))))..(((((((((((...((....))..)))))).))))).........))...)). ( -27.60) >DroSim_CAF1 49169 106 + 1 AUAAUGAAAUGCCCCAAUGGGCAAU--------------GGGCAAUGGGCAGUCAGUCGCUGCCUUCAAUUAUGCAAAACGGUUACUGCACUUUGCAUUAAUUAACAAACGGGCAAACAA .........(((((....))))).(--------------(.((...(((((((.....)))))))..(((((((((((...((....))..)))))).))))).........))...)). ( -27.60) >DroEre_CAF1 49972 113 + 1 AUAAUGAAAUGCCCCAAUGGGC-------AAUGGGUAAUGGGCAAUGGGCAGUCAGUCGCUGCCUUCAAUUAUGCAAAACGGUUACUGCACUUUGCAUUAAUUAACAAACGGGCAAACAA .........(((((...((...-------..((.(((((.......(((((((.....)))))))...))))).))....(((((.(((.....))).)))))..))...)))))..... ( -28.20) >DroYak_CAF1 49942 120 + 1 AUAAUGAAAUGCCCCAAUGGGCAAUGGGCAAUGGGUAAUGGGCAAUGGGCAGUCAGUCGCUGCCUUCAAUUAUGCAAAACGGUUACUGCACUUUGCAUUAAUUAACAAACGGGCAAACAA .........(((((....))))).((.((..((.(((((.......(((((((.....)))))))...))))).))....(((((.(((.....))).))))).........))...)). ( -29.50) >consensus AUAAUGAAAUGCCCCAAUGGGCAAU______________GGGCAAUGGGCAGUCAGUCGCUGCCUUCAAUUAUGCAAAACGGUUACUGCACUUUGCAUUAAUUAACAAACGGGCAAACAA .........(((((...((...........................(((((((.....)))))))..(((((((((((...((....))..)))))).)))))..))...)))))..... (-26.30 = -26.30 + 0.00)


| Location | 3,383,571 – 3,383,689 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.65 |
| Mean single sequence MFE | -34.25 |
| Consensus MFE | -32.73 |
| Energy contribution | -33.13 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.995387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3383571 118 + 23771897 GGCAAUGGGCAGUCAGUCGCUGCCUUCAAUUAUGCAAAACGGUUACUGCACUUUGCAUUAAUUAACAAACGGGCAAACAAAGCGAAAUUCGCGCAGCUGACACAAUUUUGCACGUGCU-- ((((..(((((((.....))))))).......(((((((..((((((((..(((((................)))))....(((.....))))))).))))....)))))))..))))-- ( -34.69) >DroSec_CAF1 51035 118 + 1 GGCAAUGGGCAGUCAGUCGCUGCCUUCAAUUAUGCAAAACGGUUACUGCACUUUGCAUUAAUUAACAAACGGGCAAACAAAGCGAAAUUCGCGCAGCUGACACAAUUUUGCACGUGCU-- ((((..(((((((.....))))))).......(((((((..((((((((..(((((................)))))....(((.....))))))).))))....)))))))..))))-- ( -34.69) >DroSim_CAF1 49195 118 + 1 GGCAAUGGGCAGUCAGUCGCUGCCUUCAAUUAUGCAAAACGGUUACUGCACUUUGCAUUAAUUAACAAACGGGCAAACAAAGCGAAAUUCGCGCAGCUGACACAAUUUUGCACGUGCU-- ((((..(((((((.....))))))).......(((((((..((((((((..(((((................)))))....(((.....))))))).))))....)))))))..))))-- ( -34.69) >DroEre_CAF1 50005 118 + 1 GGCAAUGGGCAGUCAGUCGCUGCCUUCAAUUAUGCAAAACGGUUACUGCACUUUGCAUUAAUUAACAAACGGGCAAACAAAGCGAAAUUGCCGCAGCUGACACAAUUUUGCACGUGCU-- ((((..(((((((.....))))))).......(((((((..((((((((..((((..........))))..(((((...........))))))))).))))....)))))))..))))-- ( -32.40) >DroYak_CAF1 49982 120 + 1 GGCAAUGGGCAGUCAGUCGCUGCCUUCAAUUAUGCAAAACGGUUACUGCACUUUGCAUUAAUUAACAAACGGGCAAACAAAGCGAAAUUCGCGCAGCUGACACAAUUUUGCACGUGCUUA ((((..(((((((.....))))))).......(((((((..((((((((..(((((................)))))....(((.....))))))).))))....)))))))..)))).. ( -34.79) >consensus GGCAAUGGGCAGUCAGUCGCUGCCUUCAAUUAUGCAAAACGGUUACUGCACUUUGCAUUAAUUAACAAACGGGCAAACAAAGCGAAAUUCGCGCAGCUGACACAAUUUUGCACGUGCU__ ((((..(((((((.....))))))).......(((((((..((((((((..(((((................)))))....(((.....))))))).))))....)))))))..)))).. (-32.73 = -33.13 + 0.40)



| Location | 3,383,571 – 3,383,689 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.65 |
| Mean single sequence MFE | -34.42 |
| Consensus MFE | -33.12 |
| Energy contribution | -33.32 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3383571 118 - 23771897 --AGCACGUGCAAAAUUGUGUCAGCUGCGCGAAUUUCGCUUUGUUUGCCCGUUUGUUAAUUAAUGCAAAGUGCAGUAACCGUUUUGCAUAAUUGAAGGCAGCGACUGACUGCCCAUUGCC --.(((.(((((((((.((....(((((((.......((.......))...(((((........)))))))))))).)).))))))))).......(((((.......)))))...))). ( -34.30) >DroSec_CAF1 51035 118 - 1 --AGCACGUGCAAAAUUGUGUCAGCUGCGCGAAUUUCGCUUUGUUUGCCCGUUUGUUAAUUAAUGCAAAGUGCAGUAACCGUUUUGCAUAAUUGAAGGCAGCGACUGACUGCCCAUUGCC --.(((.(((((((((.((....(((((((.......((.......))...(((((........)))))))))))).)).))))))))).......(((((.......)))))...))). ( -34.30) >DroSim_CAF1 49195 118 - 1 --AGCACGUGCAAAAUUGUGUCAGCUGCGCGAAUUUCGCUUUGUUUGCCCGUUUGUUAAUUAAUGCAAAGUGCAGUAACCGUUUUGCAUAAUUGAAGGCAGCGACUGACUGCCCAUUGCC --.(((.(((((((((.((....(((((((.......((.......))...(((((........)))))))))))).)).))))))))).......(((((.......)))))...))). ( -34.30) >DroEre_CAF1 50005 118 - 1 --AGCACGUGCAAAAUUGUGUCAGCUGCGGCAAUUUCGCUUUGUUUGCCCGUUUGUUAAUUAAUGCAAAGUGCAGUAACCGUUUUGCAUAAUUGAAGGCAGCGACUGACUGCCCAUUGCC --.(((.(((((((((.((....((((((((((...........)))))..(((((........)))))..))))).)).))))))))).......(((((.......)))))...))). ( -34.90) >DroYak_CAF1 49982 120 - 1 UAAGCACGUGCAAAAUUGUGUCAGCUGCGCGAAUUUCGCUUUGUUUGCCCGUUUGUUAAUUAAUGCAAAGUGCAGUAACCGUUUUGCAUAAUUGAAGGCAGCGACUGACUGCCCAUUGCC ...(((.(((((((((.((....(((((((.......((.......))...(((((........)))))))))))).)).))))))))).......(((((.......)))))...))). ( -34.30) >consensus __AGCACGUGCAAAAUUGUGUCAGCUGCGCGAAUUUCGCUUUGUUUGCCCGUUUGUUAAUUAAUGCAAAGUGCAGUAACCGUUUUGCAUAAUUGAAGGCAGCGACUGACUGCCCAUUGCC ...(((.(((((((((.((....(((((((.......((.......))...(((((........)))))))))))).)).))))))))).......(((((.......)))))...))). (-33.12 = -33.32 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:59 2006