
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,380,787 – 3,381,043 |
| Length | 256 |
| Max. P | 0.999937 |

| Location | 3,380,787 – 3,380,896 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.57 |
| Mean single sequence MFE | -36.04 |
| Consensus MFE | -33.30 |
| Energy contribution | -33.14 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.45 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.68 |
| SVM RNA-class probability | 0.999937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3380787 109 + 23771897 AACACCAC--AACAAGCGGCAA-------GCAGCAACUAC--AAGGGGCUGAGUGCGGCCCAUAGACAAAGUUCAACUAAGGUUGAAACCAUUUGUCGGAUUAAAUACGCGCUGCGGUAA ...(((..--..(....)....-------(((((......--...((((((....))))))...((((((.((((((....))))))....)))))).............)))))))).. ( -34.90) >DroSec_CAF1 48193 111 + 1 AACACCACUAAGCAAGCGGCAA-------GCAGCAACUAC--AAGGGGCUGAGUGCGGCCCAUAGACAAAGUUCAACUAAGGUUGAAACCAUUUGUCGGAUUAAAUACGCGCUGCGGUAA ...(((.....((.....))..-------(((((......--...((((((....))))))...((((((.((((((....))))))....)))))).............)))))))).. ( -38.30) >DroSim_CAF1 46370 109 + 1 AACGCCAC--AACAAGCGGUAA-------GCAGCAACUAC--AAGGGGCUGAGUGCGGCCCAUAGACAAAGUUCAACUAAGGUUGAAACCAUUUGUCGGAUUAAAUACGCGCUGCGGUAA ..(((...--.....)))....-------(((((......--...((((((....))))))...((((((.((((((....))))))....)))))).............)))))..... ( -36.80) >DroEre_CAF1 47140 89 + 1 -----------------------------GCAGCAACUAC--AAGGGGCUGAGUGCGGCCCAUAGACAAAGUUCAACUAAGGUUGAAACCAUUUGUCGGAUUAAAUACGCGCUGCGGUAA -----------------------------(((((......--...((((((....))))))...((((((.((((((....))))))....)))))).............)))))..... ( -33.60) >DroYak_CAF1 47065 118 + 1 AACACCAC--AACAAGCGGCAAGCAGCAAGCAGCAACUAUAGAAGGGGUUGAGUGCGGCCCAUAGACAAAGUUCAACUAAGGUUGAAACCAUUUGUCGGAUUAAAUACGCGCUGCGGUAA ...(((..--.....((.....)).....(((((...........((((((....))))))...((((((.((((((....))))))....)))))).............)))))))).. ( -36.60) >consensus AACACCAC__AACAAGCGGCAA_______GCAGCAACUAC__AAGGGGCUGAGUGCGGCCCAUAGACAAAGUUCAACUAAGGUUGAAACCAUUUGUCGGAUUAAAUACGCGCUGCGGUAA .............................(((((...........((((((....))))))...((((((.((((((....))))))....)))))).............)))))..... (-33.30 = -33.14 + -0.16)

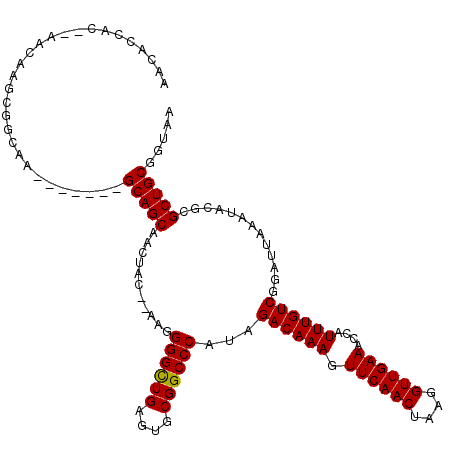
| Location | 3,380,787 – 3,380,896 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.57 |
| Mean single sequence MFE | -31.04 |
| Consensus MFE | -25.59 |
| Energy contribution | -26.24 |
| Covariance contribution | 0.65 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3380787 109 - 23771897 UUACCGCAGCGCGUAUUUAAUCCGACAAAUGGUUUCAACCUUAGUUGAACUUUGUCUAUGGGCCGCACUCAGCCCCUU--GUAGUUGCUGC-------UUGCCGCUUGUU--GUGGUGUU .(((((((((.............(((((..((((.((((....)))))))))))))...((((.(((..((((..(..--...)..)))).-------.))).)))))))--)))))).. ( -34.60) >DroSec_CAF1 48193 111 - 1 UUACCGCAGCGCGUAUUUAAUCCGACAAAUGGUUUCAACCUUAGUUGAACUUUGUCUAUGGGCCGCACUCAGCCCCUU--GUAGUUGCUGC-------UUGCCGCUUGCUUAGUGGUGUU .....(((((((...........(((((..((((.((((....)))))))))))))...((((........))))...--...).))))))-------..((((((.....))))))... ( -33.20) >DroSim_CAF1 46370 109 - 1 UUACCGCAGCGCGUAUUUAAUCCGACAAAUGGUUUCAACCUUAGUUGAACUUUGUCUAUGGGCCGCACUCAGCCCCUU--GUAGUUGCUGC-------UUACCGCUUGUU--GUGGCGUU ...((((((((((..........(((((..((((.((((....)))))))))))))..(((((.((((((((....))--).)).))).))-------))).)))..)))--)))).... ( -31.10) >DroEre_CAF1 47140 89 - 1 UUACCGCAGCGCGUAUUUAAUCCGACAAAUGGUUUCAACCUUAGUUGAACUUUGUCUAUGGGCCGCACUCAGCCCCUU--GUAGUUGCUGC----------------------------- .....(((((((...........(((((..((((.((((....)))))))))))))...((((........))))...--...).))))))----------------------------- ( -24.60) >DroYak_CAF1 47065 118 - 1 UUACCGCAGCGCGUAUUUAAUCCGACAAAUGGUUUCAACCUUAGUUGAACUUUGUCUAUGGGCCGCACUCAACCCCUUCUAUAGUUGCUGCUUGCUGCUUGCCGCUUGUU--GUGGUGUU .((((((((((((..........(((((..((((.((((....)))))))))))))...((((.(((..((((..........)))).....))).))))..)))..)))--)))))).. ( -31.70) >consensus UUACCGCAGCGCGUAUUUAAUCCGACAAAUGGUUUCAACCUUAGUUGAACUUUGUCUAUGGGCCGCACUCAGCCCCUU__GUAGUUGCUGC_______UUGCCGCUUGUU__GUGGUGUU .....(((((((...........(((((..((((.((((....)))))))))))))...((((........))))........).)))))).........(((((.......)))))... (-25.59 = -26.24 + 0.65)

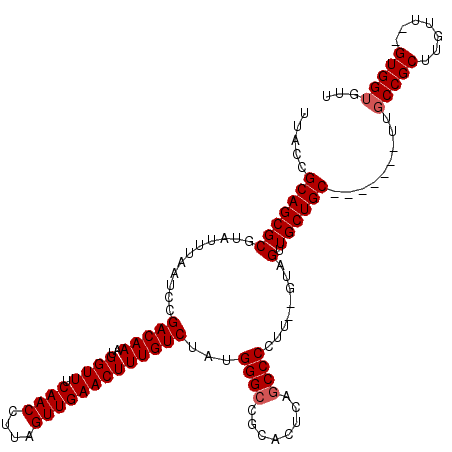

| Location | 3,380,818 – 3,380,932 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.31 |
| Mean single sequence MFE | -36.20 |
| Consensus MFE | -29.54 |
| Energy contribution | -29.58 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3380818 114 + 23771897 --AAGGGGCUGAGUGCGGCCCAUAGACAAAGUUCAACUAAGGUUGAAACCAUUUGUCGGAUUAAAUACGCGCUGCGGUAAUUAUGUGCUUCAAGCA---GCAACAACUACAC-GCCAGCA --...((((((....))))))...((((((.((((((....))))))....)))))).............(((((((((.((...((((......)---)))..)).))).)-).)))). ( -37.00) >DroSec_CAF1 48226 114 + 1 --AAGGGGCUGAGUGCGGCCCAUAGACAAAGUUCAACUAAGGUUGAAACCAUUUGUCGGAUUAAAUACGCGCUGCGGUAAUUAUGUGCUUCAAGCA---GCAACAACUACAC-GCCAGCA --...((((((....))))))...((((((.((((((....))))))....)))))).............(((((((((.((...((((......)---)))..)).))).)-).)))). ( -37.00) >DroSim_CAF1 46401 114 + 1 --AAGGGGCUGAGUGCGGCCCAUAGACAAAGUUCAACUAAGGUUGAAACCAUUUGUCGGAUUAAAUACGCGCUGCGGUAAUUAUGUGCUUCAAGCA---GCAACAACUACAC-GCCAGCA --...((((((....))))))...((((((.((((((....))))))....)))))).............(((((((((.((...((((......)---)))..)).))).)-).)))). ( -37.00) >DroEre_CAF1 47151 114 + 1 --AAGGGGCUGAGUGCGGCCCAUAGACAAAGUUCAACUAAGGUUGAAACCAUUUGUCGGAUUAAAUACGCGCUGCGGUAAUUAUGUGCUUCAAGCA---GCAACAACUACGC-GCGAGCA --...((((((....))))))...((((((.((((((....))))))....)))))).............(((((((((......))))....)))---)).........((-....)). ( -37.40) >DroYak_CAF1 47103 120 + 1 AGAAGGGGUUGAGUGCGGCCCAUAGACAAAGUUCAACUAAGGUUGAAACCAUUUGUCGGAUUAAAUACGCGCUGCGGUAAUUAUGUGCUUCAAGCAGCAGCAACAACUACACUACAAGCA ......(.(((((((..(......((((((.((((((....))))))....))))))...........(((((((((((......))))....))))).)).....)..)))).))).). ( -32.60) >consensus __AAGGGGCUGAGUGCGGCCCAUAGACAAAGUUCAACUAAGGUUGAAACCAUUUGUCGGAUUAAAUACGCGCUGCGGUAAUUAUGUGCUUCAAGCA___GCAACAACUACAC_GCCAGCA .....((((((....))))))...((((((.((((((....))))))....)))))).............(((((.(((.((...(((...........)))..)).)))...)).))). (-29.54 = -29.58 + 0.04)


| Location | 3,380,818 – 3,380,932 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.31 |
| Mean single sequence MFE | -34.76 |
| Consensus MFE | -26.74 |
| Energy contribution | -26.78 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3380818 114 - 23771897 UGCUGGC-GUGUAGUUGUUGC---UGCUUGAAGCACAUAAUUACCGCAGCGCGUAUUUAAUCCGACAAAUGGUUUCAACCUUAGUUGAACUUUGUCUAUGGGCCGCACUCAGCCCCUU-- .((((.(-(.(((((((((((---(......)))).)))))))))))))).............(((((..((((.((((....)))))))))))))...((((........))))...-- ( -36.20) >DroSec_CAF1 48226 114 - 1 UGCUGGC-GUGUAGUUGUUGC---UGCUUGAAGCACAUAAUUACCGCAGCGCGUAUUUAAUCCGACAAAUGGUUUCAACCUUAGUUGAACUUUGUCUAUGGGCCGCACUCAGCCCCUU-- .((((.(-(.(((((((((((---(......)))).)))))))))))))).............(((((..((((.((((....)))))))))))))...((((........))))...-- ( -36.20) >DroSim_CAF1 46401 114 - 1 UGCUGGC-GUGUAGUUGUUGC---UGCUUGAAGCACAUAAUUACCGCAGCGCGUAUUUAAUCCGACAAAUGGUUUCAACCUUAGUUGAACUUUGUCUAUGGGCCGCACUCAGCCCCUU-- .((((.(-(.(((((((((((---(......)))).)))))))))))))).............(((((..((((.((((....)))))))))))))...((((........))))...-- ( -36.20) >DroEre_CAF1 47151 114 - 1 UGCUCGC-GCGUAGUUGUUGC---UGCUUGAAGCACAUAAUUACCGCAGCGCGUAUUUAAUCCGACAAAUGGUUUCAACCUUAGUUGAACUUUGUCUAUGGGCCGCACUCAGCCCCUU-- .(((.((-(.(((((((((((---(......)))).)))))))))))))).............(((((..((((.((((....)))))))))))))...((((........))))...-- ( -36.20) >DroYak_CAF1 47103 120 - 1 UGCUUGUAGUGUAGUUGUUGCUGCUGCUUGAAGCACAUAAUUACCGCAGCGCGUAUUUAAUCCGACAAAUGGUUUCAACCUUAGUUGAACUUUGUCUAUGGGCCGCACUCAACCCCUUCU ...(((.(((((((((((.(.((((......))))))))))))).((.((.((((........(((((..((((.((((....))))))))))))))))).)).))))))))........ ( -29.00) >consensus UGCUGGC_GUGUAGUUGUUGC___UGCUUGAAGCACAUAAUUACCGCAGCGCGUAUUUAAUCCGACAAAUGGUUUCAACCUUAGUUGAACUUUGUCUAUGGGCCGCACUCAGCCCCUU__ .(((.((.(.(((((((((((...........))).)))))))))))))).............(((((..((((.((((....)))))))))))))...((((........))))..... (-26.74 = -26.78 + 0.04)


| Location | 3,380,932 – 3,381,043 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.80 |
| Mean single sequence MFE | -34.04 |
| Consensus MFE | -28.90 |
| Energy contribution | -28.46 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3380932 111 - 23771897 GCUAUAUAGCUUUGGCUAUAAAGGCCCAGCUGAAUCUGAAGUUCGAAACUUGUUUCGGCUUUCGG---------GGCUUUCGAAGUCGCAUUCUCAGCAUGUUUGCCAUGUUGUCUAUGU (((.((((((....))))))((((((((((((((....((((.....))))..)))))))....)---------))))))...))).((((...(((((((.....)))))))...)))) ( -34.10) >DroSec_CAF1 48340 120 - 1 GCUAUAUAGCUUUGGCUAUAAAGGCCCAGCUGAAUCUGAAGUUGGAAACUUGUUUCGGCUUUCGGUGCUUUCGAGGCUUUCGAAGUCGCAUUCUCAGCGUGUUUGCCAUGUUGUCUAUGU (((.((((((....))))))..)))((((((........))))))...........(((((.(((.(((.....)))..))))))))((((...(((((((.....)))))))...)))) ( -35.30) >DroSim_CAF1 46515 111 - 1 GCUAUAUAGCUUUGGCUAUAAAGGCCCAGCUGAAUCUGAAGUUGGAAACUUGUUUCGGCUUUCGG---------GGCUUUCGAAGUCGCAUUCUCAGCGUGUUUGCCAUGUUGUCUAUGU (((.((((((....))))))((((((((((((((....((((.....))))..)))))))....)---------))))))...))).((((...(((((((.....)))))))...)))) ( -33.70) >DroEre_CAF1 47265 111 - 1 GCUAUAUAGCUCUGGCCAUAAAGGCCCAGCCGAAUCCGAAGUUGGAAACUUGUUUCGGCUUUUGG---------GGCUCUCGAAGUCGCAUUCUCAGCAUGUUUGCCAUGUUGUCUAUGU (((....((((((((((.....)))).(((((((((((....)))).......)))))))...))---------)))).....))).((((...(((((((.....)))))))...)))) ( -34.31) >DroYak_CAF1 47223 111 - 1 GCUAUAUAGCUUCGAGUAUAAAGCACCAGCUGAGUCUGAAGUUGGAAACUUGUUUCGGCUUUCGG---------GGCUUUCGAAGUCGCAUUCUCAGCGUGUUUGCCAUGUUGUCUAUGU ........(((((((.....((((.((....(((.((((((..(....)...)))))))))...)---------)))))))))))).((((...(((((((.....)))))))...)))) ( -32.80) >consensus GCUAUAUAGCUUUGGCUAUAAAGGCCCAGCUGAAUCUGAAGUUGGAAACUUGUUUCGGCUUUCGG_________GGCUUUCGAAGUCGCAUUCUCAGCGUGUUUGCCAUGUUGUCUAUGU ......(((((..((((.....)))).)))))...((((((((((((.....))))))).))))).........((((.....))))((((...(((((((.....)))))))...)))) (-28.90 = -28.46 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:47 2006