| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,375,790 – 3,375,947 |
| Length | 157 |
| Max. P | 0.704075 |

| Location | 3,375,790 – 3,375,909 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.31 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -28.02 |
| Energy contribution | -28.22 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3375790 119 - 23771897 CUAACCGAUCCCAUCCGAUCCCUGGUUCGGUGGAAUCGACAAAAGAGCUCCACGCAUGCGCACACUUAACGAACGCUUAACGGAUGUUUCGGCUGCC-CUCCCUCCGGUUUAUAAAAAAU ....((((...((((((.......((((((((((.((.......))..)))))((....))........)))))......))))))..))))..(((-........)))........... ( -30.32) >DroSec_CAF1 42911 119 - 1 CUAACCGAUCCCAUCCGAUCCCUGGUUCGGUGGAAUCGACAAAAGAGCUCCACGCAUGCGCACACUUAACGAACGCUUAACGGAUGUUUCGGCUGCC-CUCCCUCCAGUUUAUAAAAAAU ....((((...((((((.......((((((((((.((.......))..)))))((....))........)))))......))))))..)))).....-...................... ( -29.42) >DroSim_CAF1 40644 119 - 1 CUAACCGAUCCCAUCCGAUCCCUGGUUCGGUGGAAUCGACAAAAGAGCUCCACGCAUGCGCACACUUAACGAACGCUUAACGGAUGUUUCGGCUGCC-CUCCCUCCAGUUUAUAAAAAAU ....((((...((((((.......((((((((((.((.......))..)))))((....))........)))))......))))))..)))).....-...................... ( -29.42) >DroEre_CAF1 42354 117 - 1 CCGACCGAUCCCAUCCGAUCCCUGGUUCGGUGGAAUCGACAA-AGAGCUCCACGCAUGCGCACACUUAGCGAACGCUUAACGGAUGUUUCGGCUUCC-CACCCUCCGGUUUAUAAA-AAU ..(.((((...((((((.......((((((((((.((.....-.))..)))))((....))........)))))......))))))..)))))....-.(((....))).......-... ( -30.52) >DroYak_CAF1 41862 120 - 1 CUAACAGAUUCCAUCCGAUCCCUGGUUCGGUGGAAUCGACAAAAGAGCUCCACGCAUGCGCACACUUAACGAACGCUUAACGGAUGUUUCGGCUUCCCCUCCCGCCACUUUAUAAAAAAU ......((...((((((.......((((((((((.((.......))..)))))((....))........)))))......))))))..))(((..........))).............. ( -28.32) >consensus CUAACCGAUCCCAUCCGAUCCCUGGUUCGGUGGAAUCGACAAAAGAGCUCCACGCAUGCGCACACUUAACGAACGCUUAACGGAUGUUUCGGCUGCC_CUCCCUCCAGUUUAUAAAAAAU ....((((...((((((.......((((((((((.((.......))..)))))((....))........)))))......))))))..))))............................ (-28.02 = -28.22 + 0.20)

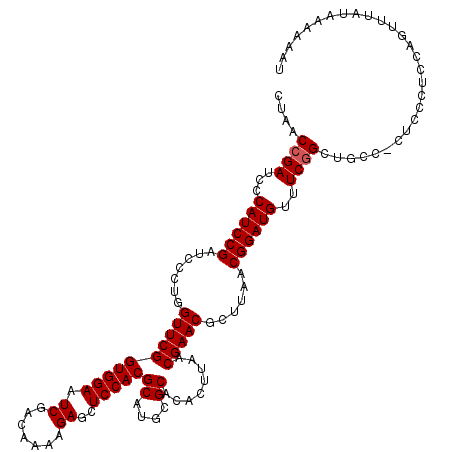
| Location | 3,375,829 – 3,375,947 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.55 |
| Mean single sequence MFE | -39.96 |
| Consensus MFE | -32.42 |
| Energy contribution | -32.70 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3375829 118 + 23771897 UUAAGCGUUCGUUAAGUGUGCGCAUGCGUGGAGCUCUUUUGUCGAUUCCACCGAACCAGGGAUCGGAUGGGAUCGGUUAGGAUCUGCCCAGAUGGAGG--GGCAUAUAGUAAGCAGUAUA ....(((..(.....)..)))((.(((.....(((((((..(((((((((((((.(....).)))).)))))))(((........)))..))..))))--))).....))).))...... ( -40.90) >DroSec_CAF1 42950 116 + 1 UUAAGCGUUCGUUAAGUGUGCGCAUGCGUGGAGCUCUUUUGUCGAUUCCACCGAACCAGGGAUCGGAUGGGAUCGGUUAGGAUCGGCCCAGAUGGAGGGAGGCAUAUA----GCAGUAUA ....((((.((.....)).)))).(((.....(((((((..(((((((((((((.(....).)))).)))))))((((......))))..))..))))..))).....----)))..... ( -37.10) >DroSim_CAF1 40683 116 + 1 UUAAGCGUUCGUUAAGUGUGCGCAUGCGUGGAGCUCUUUUGUCGAUUCCACCGAACCAGGGAUCGGAUGGGAUCGGUUAGGAUCGGCCCAGAUGGAGGGAGGCAUAUA----GCAGUAUA ....((((.((.....)).)))).(((.....(((((((..(((((((((((((.(....).)))).)))))))((((......))))..))..))))..))).....----)))..... ( -37.10) >DroEre_CAF1 42392 104 + 1 UUAAGCGUUCGCUAAGUGUGCGCAUGCGUGGAGCUCU-UUGUCGAUUCCACCGAACCAGGGAUCGGAUGGGAUCGGUCGGGAUCGG-CCAGAUGGAGGG--G------------AGUACU ....((((.(((.......))).))))(((...((((-(..(((((((((((((.(....).)))).)))))))(((((....)))-)).))..)))))--.------------..))). ( -43.80) >DroYak_CAF1 41902 110 + 1 UUAAGCGUUCGUUAAGUGUGCGCAUGCGUGGAGCUCUUUUGUCGAUUCCACCGAACCAGGGAUCGGAUGGAAUCUGUUAGGAUCGGUCCAGAUGGAGGG--GCAGA--------AGUACU ....((((.(((.......))).)))).....(((((((..(((((((((((((.(....).)))).))))))).....(((....))).))..)))))--))...--------...... ( -40.90) >consensus UUAAGCGUUCGUUAAGUGUGCGCAUGCGUGGAGCUCUUUUGUCGAUUCCACCGAACCAGGGAUCGGAUGGGAUCGGUUAGGAUCGGCCCAGAUGGAGGG_GGCAUAUA____GCAGUAUA ....((((.(((.......))).)))).....(((((((..(((((((((((((.(....).)))).))))))).....((......)).))..)))))..))................. (-32.42 = -32.70 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:35 2006