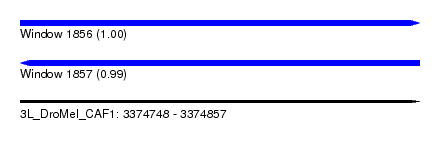
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,374,748 – 3,374,857 |
| Length | 109 |
| Max. P | 0.999966 |

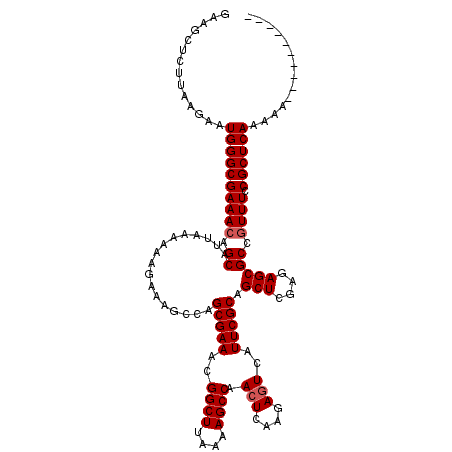
| Location | 3,374,748 – 3,374,857 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.99 |
| Mean single sequence MFE | -30.16 |
| Consensus MFE | -28.56 |
| Energy contribution | -28.96 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.98 |
| SVM RNA-class probability | 0.999966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3374748 109 + 23771897 GAAGCUCUUAAGAAUGGGCGAAACAGCAUUAAAAAAGAAAGCCAGCGAAACGGCUUAAAAGCCAACUCAAGAGUCAUUCGCAGCUCGAGAGCGCCGUUUCCGCUCAAA-A---------- ..............((((((((((.((.................(((((..((((....)))).(((....)))..))))).(((....))))).)))).))))))..-.---------- ( -30.40) >DroSec_CAF1 41889 110 + 1 GAAGCUCUUAAGAAUGGGCGAAAAAGCAUUAAAAAAGAAAGCCAGCGAAACGGCUUAAAAGCCAAAUCAAGAGUCAUUCGCAGCUCGAGAGCGCCGUUUCCGCUCAAAAA---------- ..............(((((((((..((.................(((((..((((....))))....(....)...))))).(((....)))))..))).))))))....---------- ( -24.70) >DroSim_CAF1 39617 110 + 1 GAAGCUCUUAAGAAUGGGCGAAACAGCAUUAAAAAAGAAAGCCAGCGAAACGGCUUAAAAGCCAACUCAAGAGUCAUUCGCAGCUCGAGAGCGCCGUUUCCGCUCAAAAA---------- ..............((((((((((.((.................(((((..((((....)))).(((....)))..))))).(((....))))).)))).))))))....---------- ( -30.40) >DroEre_CAF1 41276 118 + 1 GAAGCUCUUAAGAAUGGGCGAAACAGCAUUAAAAAAGAAAGCCAGCGAAACGGCUUAAAAGCCAACUCAAGAGUCAUUCGCAGCUCGAGAGCGCCGUUUCCGCUCAAAAAA--AAGAGGA ....(((((.....((((((((((.((.................(((((..((((....)))).(((....)))..))))).(((....))))).)))).)))))).....--))))).. ( -34.90) >DroYak_CAF1 40713 120 + 1 GAAGCUCUUAAGAAUGGGCGAAACAGCAUUAAAAAAGAAAGCCAGCGAAACGGCUUAAAAGCCAACUCAAGAGUCAUUCGCAGCUCGAGAGCGCCGUUUCCGCUCAAAAAAAAAAAAGAA ..............((((((((((.((.................(((((..((((....)))).(((....)))..))))).(((....))))).)))).)))))).............. ( -30.40) >consensus GAAGCUCUUAAGAAUGGGCGAAACAGCAUUAAAAAAGAAAGCCAGCGAAACGGCUUAAAAGCCAACUCAAGAGUCAUUCGCAGCUCGAGAGCGCCGUUUCCGCUCAAAAA__________ ..............((((((((((.((.................(((((..((((....)))).(((....)))..))))).(((....))))).)))).)))))).............. (-28.56 = -28.96 + 0.40)

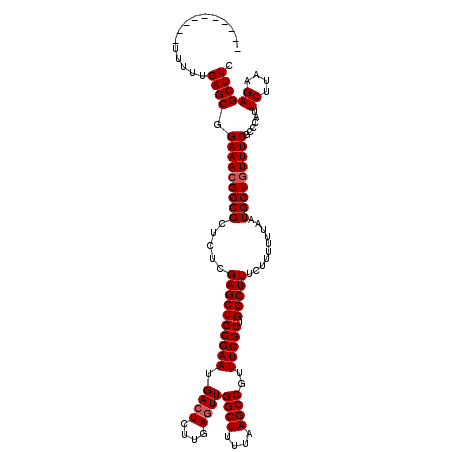
| Location | 3,374,748 – 3,374,857 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.99 |
| Mean single sequence MFE | -31.17 |
| Consensus MFE | -28.58 |
| Energy contribution | -28.98 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.994953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3374748 109 - 23771897 ----------U-UUUGAGCGGAAACGGCGCUCUCGAGCUGCGAAUGACUCUUGAGUUGGCUUUUAAGCCGUUUCGCUGGCUUUCUUUUUUAAUGCUGUUUCGCCCAUUCUUAAGAGCUUC ----------.-...((((.(((((((((.....((((((((((.((((....))))((((....))))..))))).)))))..........)))))))))......((....)))))). ( -30.56) >DroSec_CAF1 41889 110 - 1 ----------UUUUUGAGCGGAAACGGCGCUCUCGAGCUGCGAAUGACUCUUGAUUUGGCUUUUAAGCCGUUUCGCUGGCUUUCUUUUUUAAUGCUUUUUCGCCCAUUCUUAAGAGCUUC ----------........((....))..(((((..((..(((((........((..(((((....)))))..))...(((.............)))..))))).....))..)))))... ( -27.82) >DroSim_CAF1 39617 110 - 1 ----------UUUUUGAGCGGAAACGGCGCUCUCGAGCUGCGAAUGACUCUUGAGUUGGCUUUUAAGCCGUUUCGCUGGCUUUCUUUUUUAAUGCUGUUUCGCCCAUUCUUAAGAGCUUC ----------.....((((.(((((((((.....((((((((((.((((....))))((((....))))..))))).)))))..........)))))))))......((....)))))). ( -30.56) >DroEre_CAF1 41276 118 - 1 UCCUCUU--UUUUUUGAGCGGAAACGGCGCUCUCGAGCUGCGAAUGACUCUUGAGUUGGCUUUUAAGCCGUUUCGCUGGCUUUCUUUUUUAAUGCUGUUUCGCCCAUUCUUAAGAGCUUC ..(((((--.....((.(((.((((((((.....((((((((((.((((....))))((((....))))..))))).)))))..........))))))))))).)).....))))).... ( -35.46) >DroYak_CAF1 40713 120 - 1 UUCUUUUUUUUUUUUGAGCGGAAACGGCGCUCUCGAGCUGCGAAUGACUCUUGAGUUGGCUUUUAAGCCGUUUCGCUGGCUUUCUUUUUUAAUGCUGUUUCGCCCAUUCUUAAGAGCUUC ..(((((.......((.(((.((((((((.....((((((((((.((((....))))((((....))))..))))).)))))..........))))))))))).)).....))))).... ( -31.46) >consensus __________UUUUUGAGCGGAAACGGCGCUCUCGAGCUGCGAAUGACUCUUGAGUUGGCUUUUAAGCCGUUUCGCUGGCUUUCUUUUUUAAUGCUGUUUCGCCCAUUCUUAAGAGCUUC ...............((((.(((((((((.....((((((((((.((((....))))((((....))))..))))).)))))..........)))))))))......((....)))))). (-28.58 = -28.98 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:31 2006