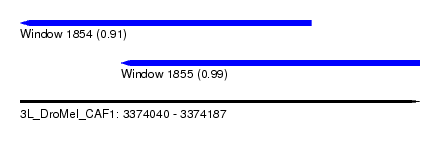
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,374,040 – 3,374,187 |
| Length | 147 |
| Max. P | 0.994666 |

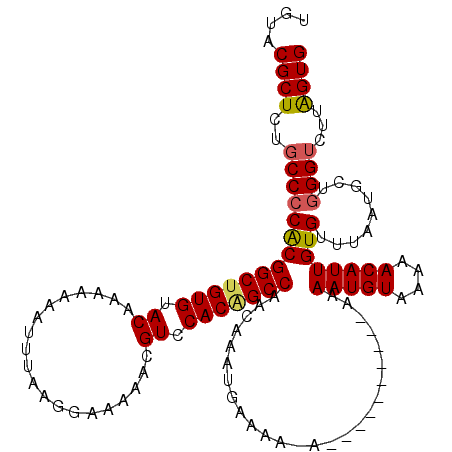
| Location | 3,374,040 – 3,374,147 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.33 |
| Mean single sequence MFE | -24.78 |
| Consensus MFE | -20.45 |
| Energy contribution | -20.61 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3374040 107 - 23771897 UGUACGCUCUGCCCCACGGCUGUGUACAAAAAAAUUUAAGGAAAAACGUCCACAGCCAACAAAUGAAAA-A----------AAAAUGUAAAAACAUUGUGUUUAAUGCUAGGG--UGGUG ....((((((((..((((((((((.((....................)).)))))))............-.----------..(((((....))))))))......)).))))--))... ( -24.45) >DroSec_CAF1 41181 110 - 1 UGUACGCUCUGCCCCACGGCUGUGUACAAAAAAAUUUAAGGAAAAACGUCCACAGCCAACAAAUGAAAAAA----------AAAAUGUAAAAACAUUGUGUUUAAUUCUGGGUCUUAGUG ....((((..(.((((.(((((((.((....................)).))))))).......(((..((----------(.(((((....)))))...)))..))))))).)..)))) ( -24.75) >DroSim_CAF1 38924 109 - 1 UGUACGCUCUGCCCCACGGCUGUGUACAAAAAAAUUUAAGGAAAAACGUCCACAGCCAACAAAUGAAAA-A----------AAAAUGUAAAAACAUUGUGUUUAAUGCUGGGUCUUAGUG ....((((..(.((((((((((((.((....................)).)))))))......((((..-.----------..(((((....)))))...))))..).)))).)..)))) ( -24.45) >DroEre_CAF1 40603 107 - 1 UGUACGCUCUGCCCCACGGCUGUGUACAAA-AAAUUUAAGGAAAACCGUCCACGGCCAGCAAAUGAAAA-A-----------AAAUGUAAAAACAUUGUGCCUAAUGCUGGGUUUUAGUG (((((((...(((....))).)))))))..-....((((((....(((....)))((((((........-.-----------.(((((....)))))........)))))).)))))).. ( -26.97) >DroYak_CAF1 40005 119 - 1 UGUGCGCUCUGCCCCGCGGCUGUGUACAAAAAAAUUUAAGGAAAACCGUCCACUGCCAACAAAUCAAAA-AAAAAAAAGGAAAAAUGUAAAAACAUUGUGUUUAAUGCUGGGUUUAAGUG ....((((..((((.(((((.(((.(((((....)))..((....)))).))).)))............-.........(((.(((((....)))))...)))...)).))))...)))) ( -23.30) >consensus UGUACGCUCUGCCCCACGGCUGUGUACAAAAAAAUUUAAGGAAAAACGUCCACAGCCAACAAAUGAAAA_A__________AAAAUGUAAAAACAUUGUGUUUAAUGCUGGGUCUUAGUG ....((((..((((((((((((((.((....................)).)))))))..........................(((((....)))))))).........))))...)))) (-20.45 = -20.61 + 0.16)


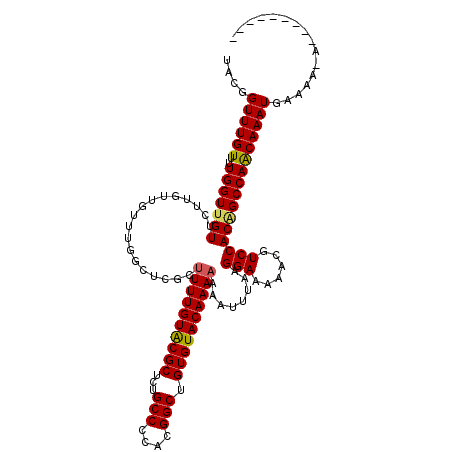
| Location | 3,374,077 – 3,374,187 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.60 |
| Mean single sequence MFE | -30.14 |
| Consensus MFE | -29.32 |
| Energy contribution | -29.28 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3374077 110 - 23771897 UACGGUUUGUUUUGGUUGUUCUUGUUGUUUGGCUCGCUUUUGUACGCUCUGCCCCACGGCUGUGUACAAAAAAAUUUAAGGAAAAACGUCCACAGCCAACAAAUGAAAA-A--------- ....((((((..(((((((..................((((((((((...(((....))).))))))))))........(((......)))))))))))))))).....-.--------- ( -30.40) >DroSec_CAF1 41220 111 - 1 UACGGUUUGUUUUGGUUGUUCUUGUUGUUUGGCUCGCUUUUGUACGCUCUGCCCCACGGCUGUGUACAAAAAAAUUUAAGGAAAAACGUCCACAGCCAACAAAUGAAAAAA--------- ....((((((..(((((((..................((((((((((...(((....))).))))))))))........(((......)))))))))))))))).......--------- ( -30.40) >DroSim_CAF1 38963 110 - 1 UACGGUUUGUUUUGGUUGUUCUUGUUGUUUGGCUCGCUUUUGUACGCUCUGCCCCACGGCUGUGUACAAAAAAAUUUAAGGAAAAACGUCCACAGCCAACAAAUGAAAA-A--------- ....((((((..(((((((..................((((((((((...(((....))).))))))))))........(((......)))))))))))))))).....-.--------- ( -30.40) >DroEre_CAF1 40641 109 - 1 UACGGUUUGUUUUGGUUGUUCUUGUUGUUUGGCUCGCUUUUGUACGCUCUGCCCCACGGCUGUGUACAAA-AAAUUUAAGGAAAACCGUCCACGGCCAGCAAAUGAAAA-A--------- ....((((((..(((((((..................((((((((((...(((....))).)))))))))-).......(((......)))))))))))))))).....-.--------- ( -29.90) >DroYak_CAF1 40045 119 - 1 UACGGUUUGUUUUGGUUGUUCUUGUUGUUUGGCUCGCUUUUGUGCGCUCUGCCCCGCGGCUGUGUACAAAAAAAUUUAAGGAAAACCGUCCACUGCCAACAAAUCAAAA-AAAAAAAAGG ..(..(((.((((((((........((((.(((....((((((((((...(((....))).))))))))))........(((......)))...)))))))))))))))-...)))..). ( -29.60) >consensus UACGGUUUGUUUUGGUUGUUCUUGUUGUUUGGCUCGCUUUUGUACGCUCUGCCCCACGGCUGUGUACAAAAAAAUUUAAGGAAAAACGUCCACAGCCAACAAAUGAAAA_A_________ ....((((((..(((((((..................((((((((((...(((....))).))))))))))........(((......))))))))))))))))................ (-29.32 = -29.28 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:29 2006