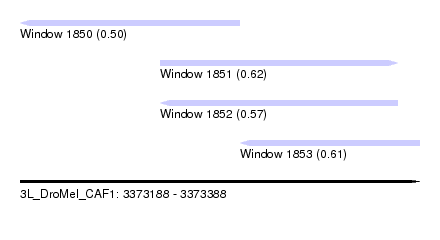
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,373,188 – 3,373,388 |
| Length | 200 |
| Max. P | 0.624763 |

| Location | 3,373,188 – 3,373,298 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 96.38 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -27.67 |
| Energy contribution | -28.18 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.93 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3373188 110 - 23771897 UACCCUAGUUUAGGGUAUUUUCGAAACGGCAGCAGCCGAAAAGUGCGACACAGCCGGCGUUGAAUGUGCCCUGAAGUUUUGUUUAUUUUCGGAGAUAAAAA-AAAAUGAAA ........((((((((((.(((((..((((.(..(((.....).))..)...))))...))))).)))))))))).((((((((........)))))))).-......... ( -29.60) >DroSec_CAF1 40320 110 - 1 UACCCUCGUUUAGGGUAUUUUCGAAACGGCAGCAGCCGAAAAGUGCGACACAGCCGGCGUUGAAUGUGCCCUGAAGUUUUGUUUAUUUUCGGAGAUAAAAA-AAAGAGAAA ....(((.((((((((((.(((((..((((.(..(((.....).))..)...))))...))))).)))))))))).((((((((........)))))))).-...)))... ( -32.80) >DroSim_CAF1 38049 111 - 1 UACCCUCGUUUAGGGUAUUUUCGAAACGGCAGCAGCCGAAAAGUGCGACACAGCCGGCGUUGAAUGUGCCCUGAAGUUUUGUUUAUUUUCGGAGAUAAAAAAAAAAAGAAA .....((.((((((((((.(((((..((((.(..(((.....).))..)...))))...))))).)))))))))).((((((((........)))))))).......)).. ( -29.70) >DroEre_CAF1 39771 110 - 1 UACCCUCGCUUAGGGUAUUUUCGAAACGGCAGCAGCCAAAAAGUGCGACACAGCCGGCGUUGAAUGUGCCCCGAAGUUUUGUUUAUUUUCGGAGAUAAAAA-AAGAAGAAA ....(((.....((((((.(((((..((((.(..(((.....).))..)...))))...))))).))))))(((((..........)))))))).......-......... ( -27.10) >consensus UACCCUCGUUUAGGGUAUUUUCGAAACGGCAGCAGCCGAAAAGUGCGACACAGCCGGCGUUGAAUGUGCCCUGAAGUUUUGUUUAUUUUCGGAGAUAAAAA_AAAAAGAAA ........((((((((((.(((((..((((.(..(((.....).))..)...))))...))))).)))))))))).((((((((........))))))))........... (-27.67 = -28.18 + 0.50)

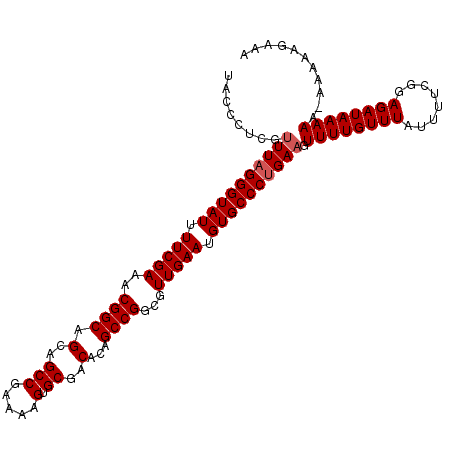
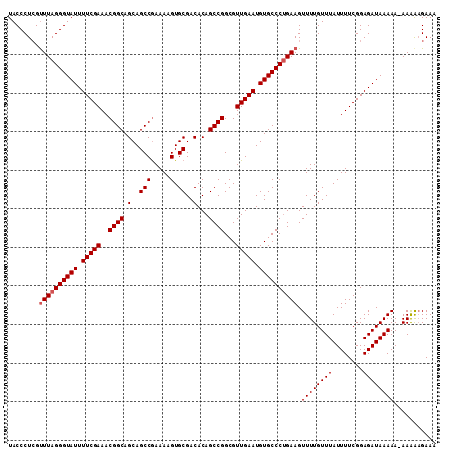
| Location | 3,373,258 – 3,373,377 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.77 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -22.30 |
| Energy contribution | -22.05 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3373258 119 + 23771897 UUCGGCUGCUGCCGUUUCGAAAAUACCCUAAACUAGGGUAUGGAAGUUUGUCCUUAAUUUUGGUUCAUAAGAUCCUGGAUUAUAUACAUAUAUAUGGGCUUAAAACCAUAUUAUGUCAA- ..((((....))))........((((((((...))))))))(((......))).(((((..((.((....)).))..)))))...(((((.((((((........)))))))))))...- ( -31.50) >DroSec_CAF1 40390 119 + 1 UUCGGCUGCUGCCGUUUCGAAAAUACCCUAAACGAGGGUAUUGAAGUUUGUCCCUAAUUUUGGUUCAUAAGAUCCUGGAUUAUAUACAUAUAUAUGGGCUUAAUACCAUAUCAUAUCAG- ..((((....))))((((((...((((((.....))))))))))))..(((...(((((..((.((....)).))..)))))...)))(((((((((........)))))).)))....- ( -28.30) >DroSim_CAF1 38120 117 + 1 UUCGGCUGCUGCCGUUUCGAAAAUACCCUAAACGAGGGUAUUGAAGUUUGUCCCUAAUUUUGGUUCACAAGAUCCUGGAUUAUAUACAUAUAUAUGGGCUUAAU--CAUAUCAUAUCAG- ..((((....)))).......((((((((.....))))))))...((..((((...((((((.....))))))...)))).....))((((((((((......)--))))).))))...- ( -27.00) >DroYak_CAF1 39149 119 + 1 UUUGGCUGCUGCCGUUUCGAAAAUACCCUAAACGAGGGUAUUCAAAUUUGUUCUUACCUUUGGU-CACUAGGUCCUUGAUUUUCUAUAUAAGUAUGUGCUAAAGAUCAUACUACUUUAGU ...(((....))).....((((((..((((..((((((((..((....))....))))))))..-...))))......))))))......((((((..(....)..))))))........ ( -23.20) >consensus UUCGGCUGCUGCCGUUUCGAAAAUACCCUAAACGAGGGUAUUGAAGUUUGUCCCUAAUUUUGGUUCACAAGAUCCUGGAUUAUAUACAUAUAUAUGGGCUUAAUACCAUAUCAUAUCAG_ ..((((....)))).......((((((((.....))))))))...((..((((...((((((.....))))))...)))).....))....((((((........))))))......... (-22.30 = -22.05 + -0.25)

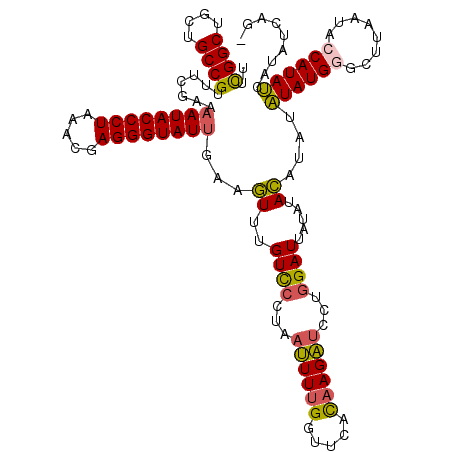
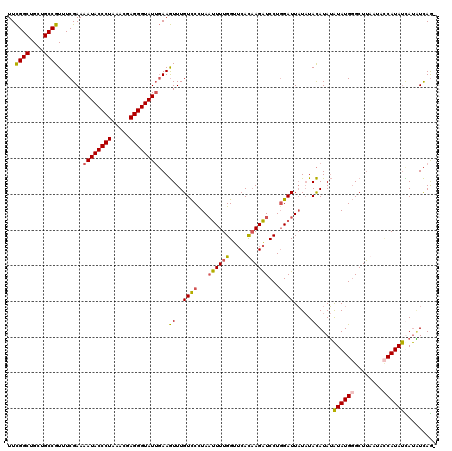
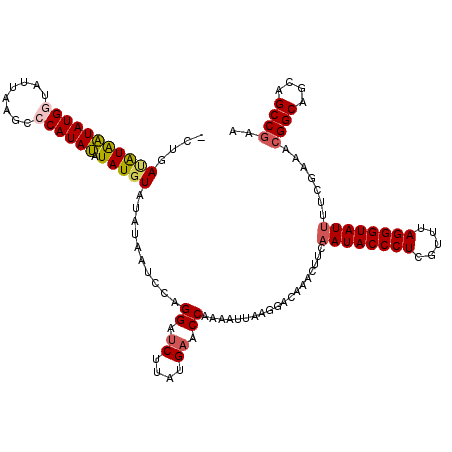
| Location | 3,373,258 – 3,373,377 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.77 |
| Mean single sequence MFE | -25.45 |
| Consensus MFE | -20.74 |
| Energy contribution | -21.30 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3373258 119 - 23771897 -UUGACAUAAUAUGGUUUUAAGCCCAUAUAUAUGUAUAUAAUCCAGGAUCUUAUGAACCAAAAUUAAGGACAAACUUCCAUACCCUAGUUUAGGGUAUUUUCGAAACGGCAGCAGCCGAA -...(((((((((((........)))))).)))))..........((.((....)).))........(((......)))((((((((...))))))))........((((....)))).. ( -28.90) >DroSec_CAF1 40390 119 - 1 -CUGAUAUGAUAUGGUAUUAAGCCCAUAUAUAUGUAUAUAAUCCAGGAUCUUAUGAACCAAAAUUAGGGACAAACUUCAAUACCCUCGUUUAGGGUAUUUUCGAAACGGCAGCAGCCGAA -(((((...((((((........))))))................((.((....)).))...)))))........(((((((((((.....))))))))...))).((((....)))).. ( -25.70) >DroSim_CAF1 38120 117 - 1 -CUGAUAUGAUAUG--AUUAAGCCCAUAUAUAUGUAUAUAAUCCAGGAUCUUGUGAACCAAAAUUAGGGACAAACUUCAAUACCCUCGUUUAGGGUAUUUUCGAAACGGCAGCAGCCGAA -...((((((((((--........))))).)))))......(((..(((.(((.....))).)))..))).....(((((((((((.....))))))))...))).((((....)))).. ( -24.40) >DroYak_CAF1 39149 119 - 1 ACUAAAGUAGUAUGAUCUUUAGCACAUACUUAUAUAGAAAAUCAAGGACCUAGUG-ACCAAAGGUAAGAACAAAUUUGAAUACCCUCGUUUAGGGUAUUUUCGAAACGGCAGCAGCCAAA .(((....((((((..........))))))....)))..........((((....-.....)))).........((((((((((((.....)))))))..)))))..(((....)))... ( -22.80) >consensus _CUGAUAUAAUAUGGUAUUAAGCCCAUAUAUAUGUAUAUAAUCCAGGAUCUUAUGAACCAAAAUUAAGGACAAACUUCAAUACCCUCGUUUAGGGUAUUUUCGAAACGGCAGCAGCCGAA ....(((((((((((........)))))).)))))..........((.((....)).))...................((((((((.....)))))))).......((((....)))).. (-20.74 = -21.30 + 0.56)

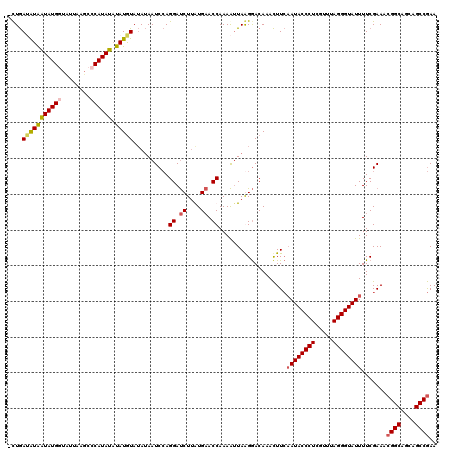
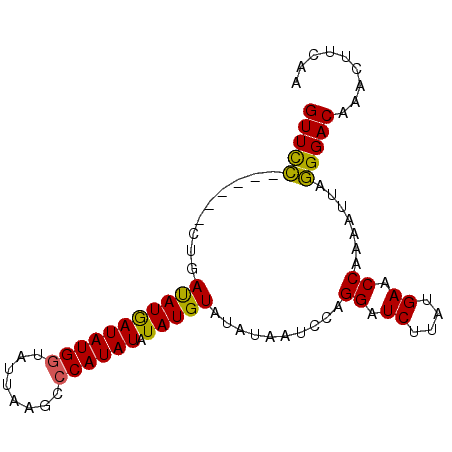
| Location | 3,373,298 – 3,373,388 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 87.12 |
| Mean single sequence MFE | -16.47 |
| Consensus MFE | -14.20 |
| Energy contribution | -13.43 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3373298 90 - 23771897 GUUUUGAAAGGUUGACAUAAUAUGGUUUUAAGCCCAUAUAUAUGUAUAUAAUCCAGGAUCUUAUGAACCAAAAUUAAGGACAAACUUCCA .........((...(((((((((((........)))))).))))).......)).((.((....)).))........(((......))). ( -15.20) >DroSec_CAF1 40430 84 - 1 GUUCC------CUGAUAUGAUAUGGUAUUAAGCCCAUAUAUAUGUAUAUAAUCCAGGAUCUUAUGAACCAAAAUUAGGGACAAACUUCAA ..(((------(((((...((((((........))))))................((.((....)).))...)))))))).......... ( -18.40) >DroSim_CAF1 38160 82 - 1 GUUCC------CUGAUAUGAUAUG--AUUAAGCCCAUAUAUAUGUAUAUAAUCCAGGAUCUUGUGAACCAAAAUUAGGGACAAACUUCAA ..(((------(((((.......(--((((.(..(((....)))..).)))))..((.((....)).))...)))))))).......... ( -15.80) >consensus GUUCC______CUGAUAUGAUAUGGUAUUAAGCCCAUAUAUAUGUAUAUAAUCCAGGAUCUUAUGAACCAAAAUUAGGGACAAACUUCAA (((((.........(((((((((((........)))))).)))))..........((.((....)).)).......)))))......... (-14.20 = -13.43 + -0.77)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:27 2006