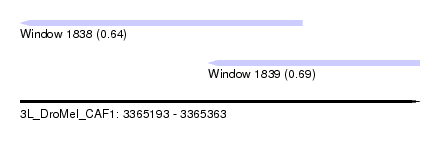
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,365,193 – 3,365,363 |
| Length | 170 |
| Max. P | 0.687792 |

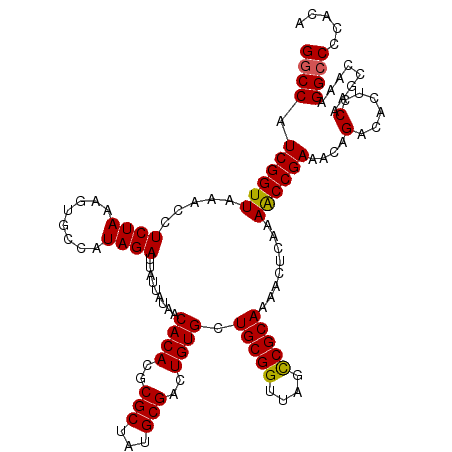
| Location | 3,365,193 – 3,365,313 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -26.44 |
| Energy contribution | -26.24 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3365193 120 - 23771897 GGCCAUCGGUUAAACCUCUAAAGUGCCAUAGAUUAUUAUAACACACGCGCUAUGCGACUGUGCUGCGGUUAGCCGCAAAACUCAAAACCGAAACAGACACUCCAAGCCAAAGGCCCCACA ((((.((((((.....((((........)))).........((((..(((...)))..)))).(((((....)))))........))))))....(......)........))))..... ( -28.30) >DroSec_CAF1 32488 120 - 1 GGCCAUCGGUUAAACCUCUAAAGUGCCAUAGAUUAUUAUAACACACGCGCUAUGCGACUGUGCUGCGGUUAGUCGCAAAACUCAAAACCGAAACAGACACUCCAAGCCAAAGGCCCCACA ((((.((((((..........(((((....................))))).((((((((.((....))))))))))........))))))....(......)........))))..... ( -28.95) >DroSim_CAF1 30167 120 - 1 GGCCAUCGGUUAAACCUCUAAAGUGCCAUAGAUUAUUAUAACACACGCGCUAUGCGACUGUGCUGCGGUUAGCCGCAAAACUCAAAACCGAAACAGACACUCCAAGCCAAAGGCCCCACA ((((.((((((.....((((........)))).........((((..(((...)))..)))).(((((....)))))........))))))....(......)........))))..... ( -28.30) >DroEre_CAF1 31188 119 - 1 GACCAUCGGUUAAACCUCUAAAGUGCCAUAGAUUAUUAUAACACACGCGCUUUGCGACUGUGCUGCGGUUAGCCGCAA-ACUCAAAGCCGAAACAGACACUCCGAGCCAAAGGCCCCACA .....((((.......(((...(((..((((....))))..))).((.((((((.....((..(((((....))))).-)).))))))))....)))....))))(((...)))...... ( -26.00) >DroYak_CAF1 31029 120 - 1 GGCCAUCGGUUAAACCUCUAAAGUGCCAUAGAUUAUUAUAACACACGCGCUAUGCGACUGUGCUGCGGUUAGCCGCAAAACUCAAAGCCGAAACAGACACUCCAAGCCAAAGGCCCCACA ((((.((((((.....((((........)))).........((((..(((...)))..)))).(((((....)))))........))))))....(......)........))))..... ( -28.30) >consensus GGCCAUCGGUUAAACCUCUAAAGUGCCAUAGAUUAUUAUAACACACGCGCUAUGCGACUGUGCUGCGGUUAGCCGCAAAACUCAAAACCGAAACAGACACUCCAAGCCAAAGGCCCCACA ((((.((((((.....((((........)))).........((((..(((...)))..)))).(((((....)))))........))))))....(......)........))))..... (-26.44 = -26.24 + -0.20)

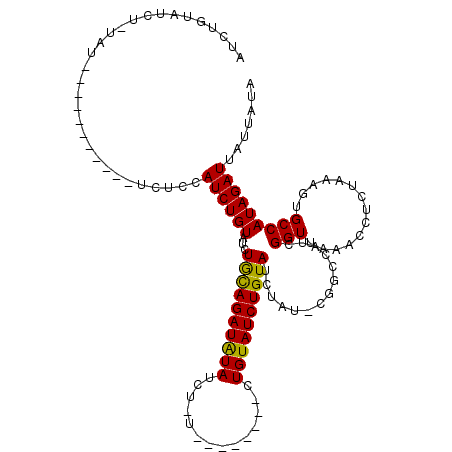
| Location | 3,365,273 – 3,365,363 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.59 |
| Mean single sequence MFE | -16.90 |
| Consensus MFE | -11.94 |
| Energy contribution | -11.45 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3365273 90 - 23771897 AUCUCUAUCUCUAU------------CUUCAUCUGUAUGUGCAGAUG------------------UAUCUGUAUCUAUCCGGCCAUCGGUUAAACCUCUAAAGUGCCAUAGAUUAUUAUA ..............------------...(((((((....)))))))------------------.......((((((..((((.(..(........)..).).)))))))))....... ( -14.50) >DroEre_CAF1 31267 91 - 1 AUCUGUA------------------UCUCCAUCUGUAUCUACAGAUAUAUCUCUG-------CUGUAUCUGUAUC----CGACCAUCGGUUAAACCUCUAAAGUGCCAUAGAUUAUUAUA .......------------------.....((((((...((((((((((......-------.)))))))))).(----((.....)))..................))))))....... ( -16.20) >DroYak_CAF1 31109 120 - 1 AACUGUAUCUGUAUCUCUAUCUGUAUCUCUAUCUGUAUCUGUAGAUAUAUCUGUAGAUAUAUCUGUAUCUGUAUCUAUGCGGCCAUCGGUUAAACCUCUAAAGUGCCAUAGAUUAUUAUA .((.((((..((((.(((((..((((.(((((........))))).))))..))))).))))..))))..))(((((((..((....((.....))......))..)))))))....... ( -20.00) >consensus AUCUGUAUCU_UAU___________UCUCCAUCUGUAUCUGCAGAUAUAUCU_U________CUGUAUCUGUAUCUAU_CGGCCAUCGGUUAAACCUCUAAAGUGCCAUAGAUUAUUAUA ..............................((((((...((((((((((..............))))))))))..............(((..............)))))))))....... (-11.94 = -11.45 + -0.49)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:13 2006