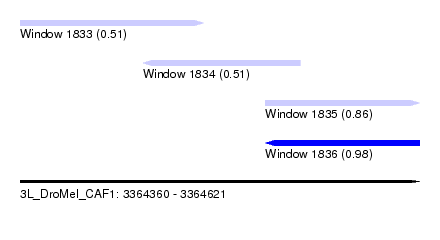
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,364,360 – 3,364,621 |
| Length | 261 |
| Max. P | 0.984225 |

| Location | 3,364,360 – 3,364,480 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.61 |
| Mean single sequence MFE | -41.27 |
| Consensus MFE | -37.47 |
| Energy contribution | -37.80 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.91 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3364360 120 + 23771897 CCCCGGGGACCAGCCCCCCAGCCCCCCAGCCCCCGCCUUCGGCCCACUCUUUCUCUACGUGCUCUCUAAUGUGCAUAUUUGGUGUGGGCUGCGUUUUGGGUUCGCCAUGUGGGCUGUCUC ....((((......))))(((((((...((.((((....((((((((.((........((((.(......).))))....)).)))))))).....))))...))...).)))))).... ( -43.40) >DroSec_CAF1 31655 111 + 1 CUCCGGGGACCAGCC---------CCCAGCCCCCGCAUUCGGCCCACUCUUUCUCUACGUGCUCUCUAAUGUGCAUAUUUGGUGUGGGCUGCGUUUUGGGUUCGCCAUGUGGGCUGUCUC .....(((((.((((---------(.((((.((((.((.((((((((.((........((((.(......).))))....)).)))))))).))..))))...))..)).)))))))))) ( -40.80) >DroSim_CAF1 29348 112 + 1 CCCCGGGGAACAGCCC--------CCCAGUCCCCGCCUUCGGCCCACUCUUUCUCUACGUGCUCUCUAAUGUGCAUAUUUGGUGUGGGCUGCGUUUUGGGUUCGCCAUGUGGGCUGUCUC .....((((.((((((--------(...(..((((....((((((((.((........((((.(......).))))....)).)))))))).....))))..).....).)))))))))) ( -39.60) >consensus CCCCGGGGACCAGCCC________CCCAGCCCCCGCCUUCGGCCCACUCUUUCUCUACGUGCUCUCUAAUGUGCAUAUUUGGUGUGGGCUGCGUUUUGGGUUCGCCAUGUGGGCUGUCUC .....((((.((((((........(((((....(((....(((((((.((........((((.(......).))))....)).))))))))))..)))))..........)))))))))) (-37.47 = -37.80 + 0.33)

| Location | 3,364,440 – 3,364,543 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.91 |
| Mean single sequence MFE | -34.92 |
| Consensus MFE | -25.94 |
| Energy contribution | -28.02 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.505780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3364440 103 - 23771897 CAUAUUUUUCGAAAAGUGUGUG-----------------GUUGAAAGUGCUGCCAAAAUGCAGUUUAAUCACCUCCGGAGGAGACAGCCCACAUGGCGAACCCAAAACGCAGCCCACACC ...............(((((.(-----------------((((...(((((((......)))))....((..((((...))))...(((.....))))).......)).)))))))))). ( -28.20) >DroSec_CAF1 31726 113 - 1 CAUAUUUUUCCAAAGGUGUGUGGUG-------GUGUGCGGUUGAAAGUGCUGCCAAAAUGCAGUUUAAUCACCUCCGGAGGAGACAGCCCACAUGGCGAACCCAAAACGCAGCCCACACC ..............((((((.(((.-------(((((.(((((.....(((((......)))))........((((...)))).)))))))))).(((.........))).))))))))) ( -37.70) >DroSim_CAF1 29420 113 - 1 CAUAUUUUUCGAAAAGUGUGUGGUG-------GUGUGCGGUUGAAAGUGCUGCCAAAAUGCAGUUUAAUCACCUCCGGAGGAGACAGCCCACAUGGCGAACCCAAAACGCAGCCCACACC ...............(((((.(((.-------(((((.(((((.....(((((......)))))........((((...)))).)))))))))).(((.........))).)))))))). ( -35.20) >DroEre_CAF1 30446 110 - 1 CAUAUUUUUCGAAAUGUGUGUGGU----------GUGCGGCUGAAAGUGCUGCCAAAAUGCAGUUUAAUCACCUCCGGAGCAGACGGCCCACAUGGCAAACCCAAAACGCAGCCCACACC (((((((....)))))))...(((----------(((.(((((...(((((((......)))))............((.((.....))))...(((.....)))..)).))))))))))) ( -32.80) >DroYak_CAF1 30309 120 - 1 CAUAUUUUUCGAAAUGUGUGUGGUGUGGUGUAGUGUGUGGUUGAAAGUGCUGCCAAAAUGCAGUUUAAUCACCUCCGGAGGAGACAGCCCACAUGGCGAACCCAAAACGCAGCCCACACC (((((((....)))))))...(((((((.((.((((((((((((....(((((......)))))))))))))....((.(....).(((.....)))....))...)))).))))))))) ( -40.70) >consensus CAUAUUUUUCGAAAAGUGUGUGGUG_______GUGUGCGGUUGAAAGUGCUGCCAAAAUGCAGUUUAAUCACCUCCGGAGGAGACAGCCCACAUGGCGAACCCAAAACGCAGCCCACACC ...............(((((.(((........(((((.(((((.....(((((......)))))....((.(((....))).)))))))))))).(((.........))).)))))))). (-25.94 = -28.02 + 2.08)

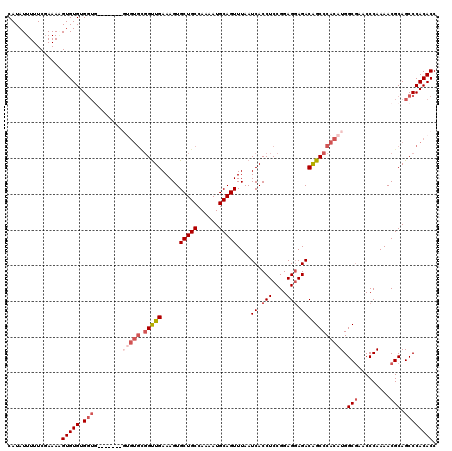
| Location | 3,364,520 – 3,364,621 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.35 |
| Mean single sequence MFE | -24.28 |
| Consensus MFE | -18.46 |
| Energy contribution | -19.06 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3364520 101 + 23771897 C-----------------CACACACUUUUCGAAAAAUAUGUGCC--CCAAGAGGGCGUGGCCAUUUCAACUUUAUGAUUUUUGUCAUAUUUCUCGAGACUUUGUGGCUUUUACAAUUCCA (-----------------((((....((((((.((((((..(((--(.....))))..(((....(((......))).....))))))))).))))))...))))).............. ( -23.90) >DroSec_CAF1 31806 110 + 1 CCGCACAC-------CACCACACACCUUUGGAAAAAUAUGUGCC--CCAAGAGGGCGUGGCCAUUUCAACUUUAUGAUUUUUGUCAUAUUUCUAGAGACUUUGUGGCU-UUACAAUUCCA ........-------..(((((...(((((((((.......(((--(.....))))(((((....(((......))).....))))).)))))))))....)))))..-........... ( -26.10) >DroSim_CAF1 29500 111 + 1 CCGCACAC-------CACCACACACUUUUCGAAAAAUAUGUGCC--CCAAGAGGGCGUGGCCAUUUCAACUUUAUGAUUUUUGUCAUAUUUCUCGAGACUUUGUGGCUUUUACAAUUCCA ........-------..(((((....((((((.((((((..(((--(.....))))..(((....(((......))).....))))))))).))))))...))))).............. ( -24.40) >DroEre_CAF1 30526 110 + 1 CCGCAC----------ACCACACACAUUUCGAAAAAUAUGUGCCCACCAAGAAGGCGUGGCCACUUCAACUUUAUGAUUUUUGUCAUAUUUCUCGAGACUUUGUGGCAUUUACAAUUCCA ......----------.(((((....((((((.(((((((((.(((((......).)))).)))...........(((....))))))))).))))))...))))).............. ( -24.00) >DroYak_CAF1 30389 119 + 1 CCACACACUACACCACACCACACACAUUUCGAAAAAUAUGUGCC-UCCAAGAAGGCGUGGCCAUUUCAACUUUAUGAUUUUUGUCAUAUUUCUCGAGACUUUGUGGCUUUUACAAUUCCA ............((((((((((((.((((....)))).)))(((-(......))))))))...((((.....((((((....))))))......))))...))))).............. ( -23.00) >consensus CCGCACAC_______CACCACACACAUUUCGAAAAAUAUGUGCC__CCAAGAGGGCGUGGCCAUUUCAACUUUAUGAUUUUUGUCAUAUUUCUCGAGACUUUGUGGCUUUUACAAUUCCA .................(((((....((((((.(((((((.(((..((.....))...)))....(((......))).......))))))).))))))...))))).............. (-18.46 = -19.06 + 0.60)


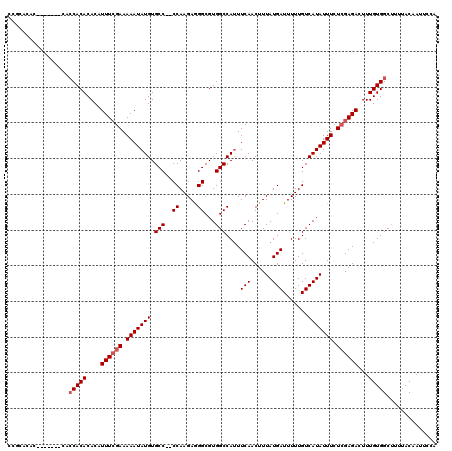
| Location | 3,364,520 – 3,364,621 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.35 |
| Mean single sequence MFE | -32.94 |
| Consensus MFE | -25.36 |
| Energy contribution | -26.16 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3364520 101 - 23771897 UGGAAUUGUAAAAGCCACAAAGUCUCGAGAAAUAUGACAAAAAUCAUAAAGUUGAAAUGGCCACGCCCUCUUGG--GGCACAUAUUUUUCGAAAAGUGUGUG-----------------G ..............(((((.....((((((((((((.......(((......))).........((((.....)--))).))))))))))))......))))-----------------) ( -29.00) >DroSec_CAF1 31806 110 - 1 UGGAAUUGUAA-AGCCACAAAGUCUCUAGAAAUAUGACAAAAAUCAUAAAGUUGAAAUGGCCACGCCCUCUUGG--GGCACAUAUUUUUCCAAAGGUGUGUGGUG-------GUGUGCGG ...........-.(((((...(((..((....)).))).....(((......)))....((((((((((.((((--((.........))))))))).))))))).-------))).)).. ( -28.10) >DroSim_CAF1 29500 111 - 1 UGGAAUUGUAAAAGCCACAAAGUCUCGAGAAAUAUGACAAAAAUCAUAAAGUUGAAAUGGCCACGCCCUCUUGG--GGCACAUAUUUUUCGAAAAGUGUGUGGUG-------GUGUGCGG ......((((...((((((.....((((((((((((.......(((......))).........((((.....)--))).))))))))))))......)))))).-------...)))). ( -34.80) >DroEre_CAF1 30526 110 - 1 UGGAAUUGUAAAUGCCACAAAGUCUCGAGAAAUAUGACAAAAAUCAUAAAGUUGAAGUGGCCACGCCUUCUUGGUGGGCACAUAUUUUUCGAAAUGUGUGUGGU----------GUGCGG ......((((...((((((..((.((((((((((((.......(((......)))....(((.((((.....))))))).)))))))))))).))...))))))----------.)))). ( -38.60) >DroYak_CAF1 30389 119 - 1 UGGAAUUGUAAAAGCCACAAAGUCUCGAGAAAUAUGACAAAAAUCAUAAAGUUGAAAUGGCCACGCCUUCUUGGA-GGCACAUAUUUUUCGAAAUGUGUGUGGUGUGGUGUAGUGUGUGG ....((((((...((((((..((.((((((((((((.......(((......))).........(((((....))-))).)))))))))))).))...))))))....))))))...... ( -34.20) >consensus UGGAAUUGUAAAAGCCACAAAGUCUCGAGAAAUAUGACAAAAAUCAUAAAGUUGAAAUGGCCACGCCCUCUUGG__GGCACAUAUUUUUCGAAAAGUGUGUGGUG_______GUGUGCGG .............((((((.....((((((((((((.......(((......)))....(((...((.....))..))).))))))))))))......))))))................ (-25.36 = -26.16 + 0.80)

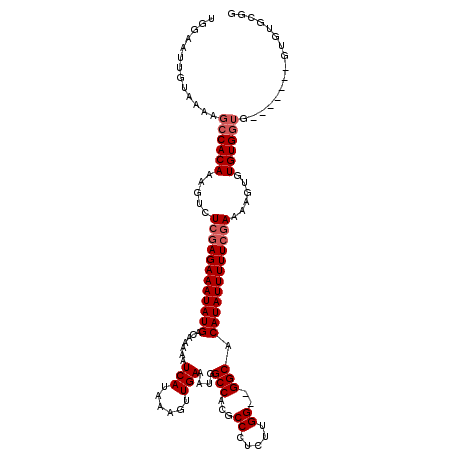
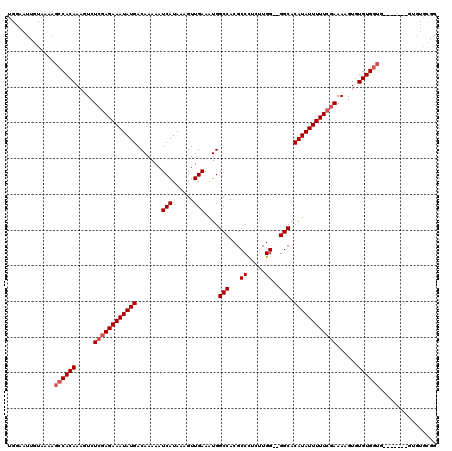
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:10 2006