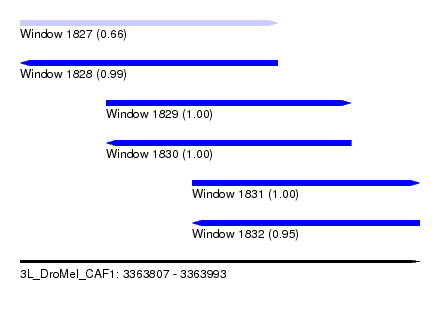
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,363,807 – 3,363,993 |
| Length | 186 |
| Max. P | 0.999946 |

| Location | 3,363,807 – 3,363,927 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -35.50 |
| Consensus MFE | -33.40 |
| Energy contribution | -33.60 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3363807 120 + 23771897 AUAUACAAUCCGAUCCACUGCGAGCCGCGCAUGCGUGACUGGUGUCCAUUGCCUUUUUGGCUUCGUUUUUGUGGAUUUUUGUGAAUGAAAUGAAUGAAAUUUCAUUCAACGUUGGCGGCG ....((((...(((((((.((((((((((....)))....((((.....)))).....))).))))....))))))).))))(((((((((.......)))))))))..((....))... ( -35.60) >DroSec_CAF1 30994 120 + 1 AUAUACAAUCCGAUCCACUGCGAGCCGCGCAUGCGUGACUGGUGUCCAUUGCCUUUUUGGCUUCAUUUUUGUGGAUUUUUGUGAAUGAAAUGAAUGAAAUUUCAUUCAACGUUGGCGGCG .......................(((((.((.((((.((....((((((.(((.....))).........))))))....))(((((((((.......))))))))).))))))))))). ( -34.90) >DroSim_CAF1 28685 120 + 1 AUAUACAAUCCGAUCCACUGCGAGCCGCGCAUGCGUGACUGGUGUCCAUUGCCUUUUUGGCUUCGUUUUUGUGGAUUUUUGUGAAUGAAAUGAAUGAAAUUUCAUUCAACGUUGGCGGCG ....((((...(((((((.((((((((((....)))....((((.....)))).....))).))))....))))))).))))(((((((((.......)))))))))..((....))... ( -35.60) >DroEre_CAF1 29764 120 + 1 AUGUACAAUCCGAUCCACUGCGAGCCGCGCAUGCGUGACUGGUGUCCAUUGCCUUUUUGGCUUCGUUUUUGUGGAUUUUUGUGAAUGAAAUGAAUGAAAUUUCAUUCAACGUUGGCGGCG ..((.((((..(((((((.((((((((((....)))....((((.....)))).....))).))))....)))))))....((((((((((.......))))))))))..))))...)). ( -36.10) >DroYak_CAF1 29559 120 + 1 AUGUACAAUCCGAUCCACUGCGAACCGCGCAUGCGUGACUGGUGUCCAUUGCCUUUUUGGCUUCGUUUUGGUGGAUUUUUGUGAAUGAAAUGAAUGAAAUUUCAUUCAACGUUGGCGGCG ..((.((((..(((((((((((((.((((....)))).............(((.....))))))))...))))))))....((((((((((.......))))))))))..))))...)). ( -35.30) >consensus AUAUACAAUCCGAUCCACUGCGAGCCGCGCAUGCGUGACUGGUGUCCAUUGCCUUUUUGGCUUCGUUUUUGUGGAUUUUUGUGAAUGAAAUGAAUGAAAUUUCAUUCAACGUUGGCGGCG .......................(((((.((.((((.((....((((((.(((.....))).........))))))....))(((((((((.......))))))))).))))))))))). (-33.40 = -33.60 + 0.20)


| Location | 3,363,807 – 3,363,927 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -30.04 |
| Consensus MFE | -29.76 |
| Energy contribution | -29.60 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3363807 120 - 23771897 CGCCGCCAACGUUGAAUGAAAUUUCAUUCAUUUCAUUCACAAAAAUCCACAAAAACGAAGCCAAAAAGGCAAUGGACACCAGUCACGCAUGCGCGGCUCGCAGUGGAUCGGAUUGUAUAU ..(((.....((.(((((((((.......)))))))))))....((((((.....((..(((.....)))....(((....))).))..((((.....)))))))))))))......... ( -30.10) >DroSec_CAF1 30994 120 - 1 CGCCGCCAACGUUGAAUGAAAUUUCAUUCAUUUCAUUCACAAAAAUCCACAAAAAUGAAGCCAAAAAGGCAAUGGACACCAGUCACGCAUGCGCGGCUCGCAGUGGAUCGGAUUGUAUAU ..(((.....((.(((((((((.......)))))))))))....((((((.........(((.....)))..........((((.((....)).))))....)))))))))......... ( -29.80) >DroSim_CAF1 28685 120 - 1 CGCCGCCAACGUUGAAUGAAAUUUCAUUCAUUUCAUUCACAAAAAUCCACAAAAACGAAGCCAAAAAGGCAAUGGACACCAGUCACGCAUGCGCGGCUCGCAGUGGAUCGGAUUGUAUAU ..(((.....((.(((((((((.......)))))))))))....((((((.....((..(((.....)))....(((....))).))..((((.....)))))))))))))......... ( -30.10) >DroEre_CAF1 29764 120 - 1 CGCCGCCAACGUUGAAUGAAAUUUCAUUCAUUUCAUUCACAAAAAUCCACAAAAACGAAGCCAAAAAGGCAAUGGACACCAGUCACGCAUGCGCGGCUCGCAGUGGAUCGGAUUGUACAU ..(((.....((.(((((((((.......)))))))))))....((((((.....((..(((.....)))....(((....))).))..((((.....)))))))))))))......... ( -30.10) >DroYak_CAF1 29559 120 - 1 CGCCGCCAACGUUGAAUGAAAUUUCAUUCAUUUCAUUCACAAAAAUCCACCAAAACGAAGCCAAAAAGGCAAUGGACACCAGUCACGCAUGCGCGGUUCGCAGUGGAUCGGAUUGUACAU ..(((.....((.(((((((((.......)))))))))))....((((((.....(((((((.....)))....(((....))).(((....))).))))..)))))))))......... ( -30.10) >consensus CGCCGCCAACGUUGAAUGAAAUUUCAUUCAUUUCAUUCACAAAAAUCCACAAAAACGAAGCCAAAAAGGCAAUGGACACCAGUCACGCAUGCGCGGCUCGCAGUGGAUCGGAUUGUAUAU ..(((.....((.(((((((((.......)))))))))))....((((((.....((..(((.....)))....(((....))).))..((((.....)))))))))))))......... (-29.76 = -29.60 + -0.16)


| Location | 3,363,847 – 3,363,961 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.77 |
| Mean single sequence MFE | -44.98 |
| Consensus MFE | -41.10 |
| Energy contribution | -41.10 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.49 |
| SVM RNA-class probability | 0.999908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3363847 114 + 23771897 GGUGUCCAUUGCCUUUUUGGCUUCGUUUUUGUGGAUUUUUGUGAAUGAAAUGAAUGAAAUUUCAUUCAACGUUGGCGGCGCGCGUCUCGCUGCGUUGGCGUCGGC------AGCGCCGUC ...((((((.(((.....))).........)))))).....((((((((((.......)))))))))).....((((((((..(((.((((.....))))..)))------.)))))))) ( -42.20) >DroSec_CAF1 31034 120 + 1 GGUGUCCAUUGCCUUUUUGGCUUCAUUUUUGUGGAUUUUUGUGAAUGAAAUGAAUGAAAUUUCAUUCAACGUUGGCGGCGCGCGUCUCGCUGCGUUGGCGUCGGCAGAGGCAGCGCCGUC ...((((((.(((.....))).........)))))).....((((((((((.......)))))))))).....((((((((..(((((((((((....)).)))).))))).)))))))) ( -49.50) >DroSim_CAF1 28725 120 + 1 GGUGUCCAUUGCCUUUUUGGCUUCGUUUUUGUGGAUUUUUGUGAAUGAAAUGAAUGAAAUUUCAUUCAACGUUGGCGGCGCGCGUCUCGCUGCGUUGGCGUCGGCAGAGGCAGCGCCGUC ...((((((.(((.....))).........)))))).....((((((((((.......)))))))))).....((((((((..(((((((((((....)).)))).))))).)))))))) ( -49.50) >DroEre_CAF1 29804 114 + 1 GGUGUCCAUUGCCUUUUUGGCUUCGUUUUUGUGGAUUUUUGUGAAUGAAAUGAAUGAAAUUUCAUUCAACGUUGGCGGCGCGCGUCUUGCUGCGUUGGCGUCGGC------AGCGCCGUC (((((((((.(((.....))).........)))))).....((((((((((.......))))))))))..((((.((((((((((......))))..)))))).)------))))))... ( -41.80) >DroYak_CAF1 29599 114 + 1 GGUGUCCAUUGCCUUUUUGGCUUCGUUUUGGUGGAUUUUUGUGAAUGAAAUGAAUGAAAUUUCAUUCAACGUUGGCGGCGCGCGUCUUGCUGCGUUGGCGUCGGC------AGCGCCGUC (((((((((((((.....)))........))))))).....((((((((((.......))))))))))..((((.((((((((((......))))..)))))).)------))))))... ( -41.90) >consensus GGUGUCCAUUGCCUUUUUGGCUUCGUUUUUGUGGAUUUUUGUGAAUGAAAUGAAUGAAAUUUCAUUCAACGUUGGCGGCGCGCGUCUCGCUGCGUUGGCGUCGGC______AGCGCCGUC ...((((((.(((.....))).........)))))).....((((((((((.......)))))))))).....((((((((.......((((((....)).)))).......)))))))) (-41.10 = -41.10 + 0.00)

| Location | 3,363,847 – 3,363,961 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.77 |
| Mean single sequence MFE | -32.74 |
| Consensus MFE | -30.60 |
| Energy contribution | -30.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.55 |
| SVM RNA-class probability | 0.999919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3363847 114 - 23771897 GACGGCGCU------GCCGACGCCAACGCAGCGAGACGCGCGCCGCCAACGUUGAAUGAAAUUUCAUUCAUUUCAUUCACAAAAAUCCACAAAAACGAAGCCAAAAAGGCAAUGGACACC ..((((((.------((...(((.......)))....))))))))(((..((.(((((((((.......)))))))))))...................(((.....)))..)))..... ( -31.70) >DroSec_CAF1 31034 120 - 1 GACGGCGCUGCCUCUGCCGACGCCAACGCAGCGAGACGCGCGCCGCCAACGUUGAAUGAAAUUUCAUUCAUUUCAUUCACAAAAAUCCACAAAAAUGAAGCCAAAAAGGCAAUGGACACC ..((((((.((.(((((.(.((....))).)).))).))))))))(((..((.(((((((((.......)))))))))))...................(((.....)))..)))..... ( -35.70) >DroSim_CAF1 28725 120 - 1 GACGGCGCUGCCUCUGCCGACGCCAACGCAGCGAGACGCGCGCCGCCAACGUUGAAUGAAAUUUCAUUCAUUUCAUUCACAAAAAUCCACAAAAACGAAGCCAAAAAGGCAAUGGACACC ..((((((.((.(((((.(.((....))).)).))).))))))))(((..((.(((((((((.......)))))))))))...................(((.....)))..)))..... ( -35.70) >DroEre_CAF1 29804 114 - 1 GACGGCGCU------GCCGACGCCAACGCAGCAAGACGCGCGCCGCCAACGUUGAAUGAAAUUUCAUUCAUUUCAUUCACAAAAAUCCACAAAAACGAAGCCAAAAAGGCAAUGGACACC ..((((((.------......((....)).((.....))))))))(((..((.(((((((((.......)))))))))))...................(((.....)))..)))..... ( -30.30) >DroYak_CAF1 29599 114 - 1 GACGGCGCU------GCCGACGCCAACGCAGCAAGACGCGCGCCGCCAACGUUGAAUGAAAUUUCAUUCAUUUCAUUCACAAAAAUCCACCAAAACGAAGCCAAAAAGGCAAUGGACACC ..((((((.------......((....)).((.....))))))))(((..((.(((((((((.......)))))))))))...................(((.....)))..)))..... ( -30.30) >consensus GACGGCGCU______GCCGACGCCAACGCAGCGAGACGCGCGCCGCCAACGUUGAAUGAAAUUUCAUUCAUUUCAUUCACAAAAAUCCACAAAAACGAAGCCAAAAAGGCAAUGGACACC ..((((((.............((....)).((.....))))))))(((..((.(((((((((.......)))))))))))...................(((.....)))..)))..... (-30.60 = -30.60 + -0.00)


| Location | 3,363,887 – 3,363,993 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.12 |
| Mean single sequence MFE | -51.80 |
| Consensus MFE | -45.96 |
| Energy contribution | -46.56 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.89 |
| SVM decision value | 4.75 |
| SVM RNA-class probability | 0.999946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3363887 106 + 23771897 GUGAAUGAAAUGAAUGAAAUUUCAUUCAACGUUGGCGGCGCGCGUCUCGCUGCGUUGGCGUCGGC------AGCGCCGUCGACGCCCAAAUCGGGAGAGGAUGGGGAGAGGA-------- .((((((((((.......)))))))))).((((((((((((..(((.((((.....))))..)))------.))))))))))))((((..((....))...)))).......-------- ( -49.80) >DroSec_CAF1 31074 120 + 1 GUGAAUGAAAUGAAUGAAAUUUCAUUCAACGUUGGCGGCGCGCGUCUCGCUGCGUUGGCGUCGGCAGAGGCAGCGCCGUCGACGCCCAAAUCGGGAGAGGAUGGGGAGAGGAGCGUGGGG .((((((((((.......)))))))))).((((((((((((..(((((((((((....)).)))).))))).))))))))))))((((..((....))...))))............... ( -57.10) >DroSim_CAF1 28765 120 + 1 GUGAAUGAAAUGAAUGAAAUUUCAUUCAACGUUGGCGGCGCGCGUCUCGCUGCGUUGGCGUCGGCAGAGGCAGCGCCGUCGACGCCCAAAUCGGGAGAGGAUGGGGAGAGGAGCGUGGGG .((((((((((.......)))))))))).((((((((((((..(((((((((((....)).)))).))))).))))))))))))((((..((....))...))))............... ( -57.10) >DroEre_CAF1 29844 114 + 1 GUGAAUGAAAUGAAUGAAAUUUCAUUCAACGUUGGCGGCGCGCGUCUUGCUGCGUUGGCGUCGGC------AGCGCCGUCGCCGCCCAAAUCGGGAGAGGAUGGAGAGAGGAGCGUGGGG .((((((((((.......))))))))))((((((((((((.((((..(((((((....)).))))------))))))))))))..(((..((....))...))).......))))).... ( -47.60) >DroYak_CAF1 29639 112 + 1 GUGAAUGAAAUGAAUGAAAUUUCAUUCAACGUUGGCGGCGCGCGUCUUGCUGCGUUGGCGUCGGC------AGCGCCGUCGACGCCCAAAUCGGGAGCGGAUGGGGAGAGGAGUGUGG-- .((((((((((.......)))))))))).((((((((((((..(((.((((.....))))..)))------.))))))))))))((((..(((....))).)))).............-- ( -47.40) >consensus GUGAAUGAAAUGAAUGAAAUUUCAUUCAACGUUGGCGGCGCGCGUCUCGCUGCGUUGGCGUCGGC______AGCGCCGUCGACGCCCAAAUCGGGAGAGGAUGGGGAGAGGAGCGUGGGG .((((((((((.......)))))))))).((((((((((((.......((((((....)).)))).......))))))))))))((((..((....))...))))............... (-45.96 = -46.56 + 0.60)


| Location | 3,363,887 – 3,363,993 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.12 |
| Mean single sequence MFE | -33.64 |
| Consensus MFE | -29.12 |
| Energy contribution | -29.36 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3363887 106 - 23771897 --------UCCUCUCCCCAUCCUCUCCCGAUUUGGGCGUCGACGGCGCU------GCCGACGCCAACGCAGCGAGACGCGCGCCGCCAACGUUGAAUGAAAUUUCAUUCAUUUCAUUCAC --------.......((((...((....))..))))(((.(.((((((.------((...(((.......)))....)))))))).).))).((((((((((.......)))))))))). ( -31.90) >DroSec_CAF1 31074 120 - 1 CCCCACGCUCCUCUCCCCAUCCUCUCCCGAUUUGGGCGUCGACGGCGCUGCCUCUGCCGACGCCAACGCAGCGAGACGCGCGCCGCCAACGUUGAAUGAAAUUUCAUUCAUUUCAUUCAC ...............((((...((....))..))))(((.(.((((((.((.(((((.(.((....))).)).))).)))))))).).))).((((((((((.......)))))))))). ( -35.90) >DroSim_CAF1 28765 120 - 1 CCCCACGCUCCUCUCCCCAUCCUCUCCCGAUUUGGGCGUCGACGGCGCUGCCUCUGCCGACGCCAACGCAGCGAGACGCGCGCCGCCAACGUUGAAUGAAAUUUCAUUCAUUUCAUUCAC ...............((((...((....))..))))(((.(.((((((.((.(((((.(.((....))).)).))).)))))))).).))).((((((((((.......)))))))))). ( -35.90) >DroEre_CAF1 29844 114 - 1 CCCCACGCUCCUCUCUCCAUCCUCUCCCGAUUUGGGCGGCGACGGCGCU------GCCGACGCCAACGCAGCAAGACGCGCGCCGCCAACGUUGAAUGAAAUUUCAUUCAUUUCAUUCAC ..................................(((((((..(((((.------...).))))..(((........)))))))))).....((((((((((.......)))))))))). ( -34.00) >DroYak_CAF1 29639 112 - 1 --CCACACUCCUCUCCCCAUCCGCUCCCGAUUUGGGCGUCGACGGCGCU------GCCGACGCCAACGCAGCAAGACGCGCGCCGCCAACGUUGAAUGAAAUUUCAUUCAUUUCAUUCAC --.............((((..((....))...))))(((.(.((((((.------......((....)).((.....)))))))).).))).((((((((((.......)))))))))). ( -30.50) >consensus CCCCACGCUCCUCUCCCCAUCCUCUCCCGAUUUGGGCGUCGACGGCGCU______GCCGACGCCAACGCAGCGAGACGCGCGCCGCCAACGUUGAAUGAAAUUUCAUUCAUUUCAUUCAC ...............((((...((....))..))))(((.(.((((((.............((....)).((.....)))))))).).))).((((((((((.......)))))))))). (-29.12 = -29.36 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:07 2006