
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,363,593 – 3,363,705 |
| Length | 112 |
| Max. P | 0.776175 |

| Location | 3,363,593 – 3,363,705 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.01 |
| Mean single sequence MFE | -34.42 |
| Consensus MFE | -25.98 |
| Energy contribution | -26.02 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3363593 112 + 23771897 AAUAAAGGCAGCGCGACAAACGAUCUCGAGACG-GCGAAAUAUCUUGUCGCCAUAAUUGGAAUUUAAUACUUCCGGUUGGGCAACAACGUUUGCAAGC-------CGCGAGACGCCCAGA ......(((....((.....)).(((((...((-((.....((.((((.(((.(((((((((........)))))))))))).)))).))......))-------))))))).))).... ( -33.10) >DroSec_CAF1 30771 119 + 1 AAUAAAGGCAGCGCGACAAACGAUCUCGAGACG-GCGAAAUAUCUUGUCGCCAUAAUUGGAAUUUAAUACUUCCGGUUGGGCAACAACGUUUGCGAGCCGCGAGCCGCGAGACGCCCAGA ......(((.((.((.((((((..........(-((((......)))))(((.(((((((((........)))))))))))).....)))))))).))((((...))))....))).... ( -38.30) >DroSim_CAF1 28469 112 + 1 AAUAAAGGCAGCGCGACAAACGAUCUCGAGACG-GCGAAAUAUCUUGUCGCCAUAAUUGGAAUUUAAUACUUCCGGUUGGGCAACAACGUUUGCGAGC-------CGCGAGACGCCCAGA ......(((.((.((.((((((..........(-((((......)))))(((.(((((((((........)))))))))))).....)))))))).))-------(....)..))).... ( -34.90) >DroEre_CAF1 29551 106 + 1 AAUAAAGGCAGCGCGACAAACGAUCUCGAGACGGGCAAAAUAUCUUGUCGCCAUAAUUGGAAUUUAAUACUUCCGGUUGGGCAACAACGUUUGC--------------GAGACGCCCAGA ......(((..(.((.((((((.......((((((........))))))(((.(((((((((........)))))))))))).....)))))))--------------).)..))).... ( -33.70) >DroYak_CAF1 29362 105 + 1 AAUAAAGGCAGCGCGACAAACGAUCUCGAGACG-GCGAAAUAUCUUGUCGCCAUAAUUGGAAUUUAAUACUUCCGGUUGGGCAACAACGUUUGC--------------GAGACGCCCAGA ......(((..(.((.((((((..........(-((((......)))))(((.(((((((((........)))))))))))).....)))))))--------------).)..))).... ( -32.10) >consensus AAUAAAGGCAGCGCGACAAACGAUCUCGAGACG_GCGAAAUAUCUUGUCGCCAUAAUUGGAAUUUAAUACUUCCGGUUGGGCAACAACGUUUGC_AGC_______CGCGAGACGCCCAGA ......(((....((.....)).((((.......(((((.....((((.(((.(((((((((........)))))))))))).))))..)))))..((........)))))).))).... (-25.98 = -26.02 + 0.04)

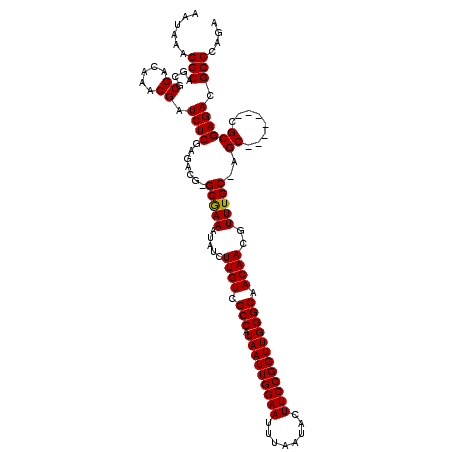
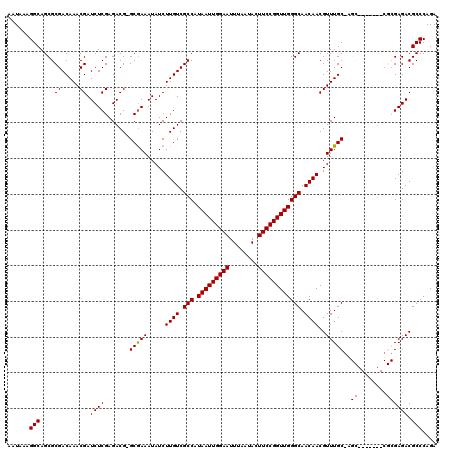
| Location | 3,363,593 – 3,363,705 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.01 |
| Mean single sequence MFE | -34.68 |
| Consensus MFE | -27.76 |
| Energy contribution | -28.80 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.776175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3363593 112 - 23771897 UCUGGGCGUCUCGCG-------GCUUGCAAACGUUGUUGCCCAACCGGAAGUAUUAAAUUCCAAUUAUGGCGACAAGAUAUUUCGC-CGUCUCGAGAUCGUUUGUCGCGCUGCCUUUAUU ...(((((...((((-------((..((.....((((((((.....((((........))))......))))))))...((((((.-.....)))))).))..)))))).)))))..... ( -35.20) >DroSec_CAF1 30771 119 - 1 UCUGGGCGUCUCGCGGCUCGCGGCUCGCAAACGUUGUUGCCCAACCGGAAGUAUUAAAUUCCAAUUAUGGCGACAAGAUAUUUCGC-CGUCUCGAGAUCGUUUGUCGCGCUGCCUUUAUU ...(((((...((((((..((((((((...(((((((((((.....((((........))))......))))))))((....))..-)))..)))).))))..)))))).)))))..... ( -41.40) >DroSim_CAF1 28469 112 - 1 UCUGGGCGUCUCGCG-------GCUCGCAAACGUUGUUGCCCAACCGGAAGUAUUAAAUUCCAAUUAUGGCGACAAGAUAUUUCGC-CGUCUCGAGAUCGUUUGUCGCGCUGCCUUUAUU ............(((-------((.((((((((((((((((.....((((........))))......))))))))...((((((.-.....)))))))))))).)).)))))....... ( -34.80) >DroEre_CAF1 29551 106 - 1 UCUGGGCGUCUC--------------GCAAACGUUGUUGCCCAACCGGAAGUAUUAAAUUCCAAUUAUGGCGACAAGAUAUUUUGCCCGUCUCGAGAUCGUUUGUCGCGCUGCCUUUAUU ...(((((.(.(--------------(((((((((((((((.....((((........))))......))))))))...((((((.......)))))))))))).)).).)))))..... ( -29.70) >DroYak_CAF1 29362 105 - 1 UCUGGGCGUCUC--------------GCAAACGUUGUUGCCCAACCGGAAGUAUUAAAUUCCAAUUAUGGCGACAAGAUAUUUCGC-CGUCUCGAGAUCGUUUGUCGCGCUGCCUUUAUU ...(((((.(.(--------------(((((((((((((((.....((((........))))......))))))))...((((((.-.....)))))))))))).)).).)))))..... ( -32.30) >consensus UCUGGGCGUCUCGCG_______GCU_GCAAACGUUGUUGCCCAACCGGAAGUAUUAAAUUCCAAUUAUGGCGACAAGAUAUUUCGC_CGUCUCGAGAUCGUUUGUCGCGCUGCCUUUAUU ...(((((...((((...........(((((((((((((((.....((((........))))......))))))))...((((((.......))))))))))))))))).)))))..... (-27.76 = -28.80 + 1.04)

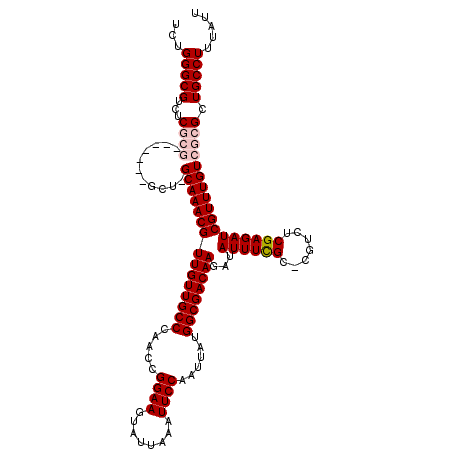
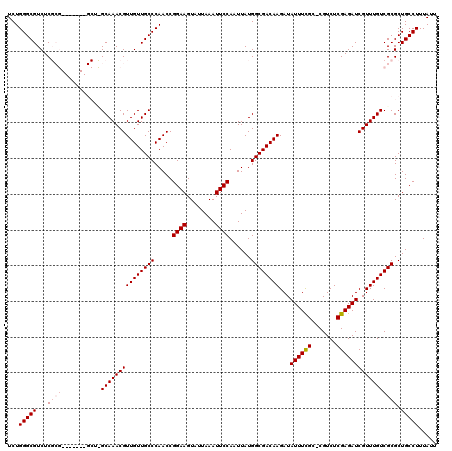
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:01 2006