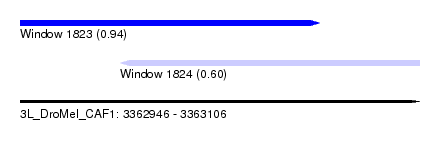
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,362,946 – 3,363,106 |
| Length | 160 |
| Max. P | 0.938599 |

| Location | 3,362,946 – 3,363,066 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.75 |
| Mean single sequence MFE | -41.26 |
| Consensus MFE | -37.60 |
| Energy contribution | -37.56 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
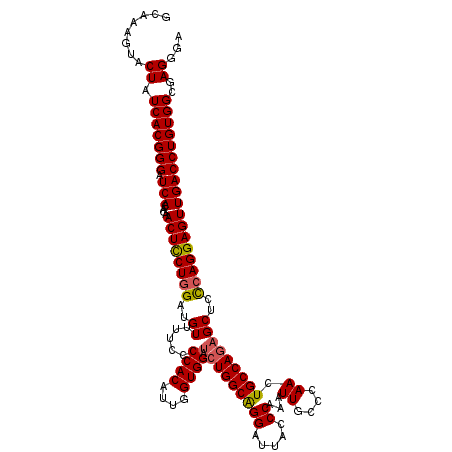
>3L_DroMel_CAF1 3362946 120 + 23771897 ACGAAGUACUAUCACGGGAUCACAGAACUCCUUGAUGUUUUCCCCACUUUGGUGGAUCUGGCAGGAUUACCCAAAUUGGACAAAUGCCAGAGCUCUCAGGAGUUGACCUGUGGCGAGGGA ........((.(((((((.(((....((((((.((........((((....)))).(((((((.......((.....)).....)))))))....))))))))))))))))))..))... ( -37.50) >DroSec_CAF1 30104 120 + 1 GCAAAGUACUAUCACGGGAUCACAGAACUCCUGGAUGUUUUCCCCACAUUGGUGGAUCUGGCAGGAUUACCCAAAUUGCCCAAGUGCCAGAGCUCCCAGGAGUUGACCUGUGGCGAGGGA ........((.(((((((.(((....((((((((..((..(((((.....)).)))((((((.((.....)).....((....))))))))))..))))))))))))))))))..))... ( -42.10) >DroSim_CAF1 27805 120 + 1 GCAAAAUACUAUCACGGGAUCACAGAACUCCUGGAUGUUUUCCCCACAUUGGUGGAUAUGGCAGGAUUACCCAAAUUGCCCAAGUGCCAGAGCUCCCAGGAGUUGACCUGUGGCGAGGGA ........((.(((((((.(((....((((((((..(((((((((.....)).)))..((((.((.....)).....((....))))))))))..))))))))))))))))))..))... ( -39.50) >DroEre_CAF1 28902 120 + 1 ACCAGAUACUAUCACGGGAUCACAGAACUUCUGGAUGUUUUUCCCACUUUGGUGGACCUGGCGGGAUUACCCAAGUUGCCCAACUGCCAGAGCUCCCAGGAGUUGACCUGUGGCGAGGGA ........((.(((((((.(((....((((((((..(((....((((....))))..((((((((....))).((((....))))))))))))..))))))))))))))))))..))... ( -44.50) >DroYak_CAF1 28687 120 + 1 GCAAGGUACUAUCACGGGAUCACAGAACUUCUGGAUGUUUUUCCCACCUUGGUGGAUCUGGCGGGAUUACCCAAAUUGCCCAACUGCCAGAGCUCCCAGGAGUUGACCUGUGGCGAGGGA ........((.(((((((.(((....((((((((..((.....((((....)))).((((((((...................))))))))))..))))))))))))))))))..))... ( -42.71) >consensus GCAAAGUACUAUCACGGGAUCACAGAACUCCUGGAUGUUUUCCCCACAUUGGUGGAUCUGGCAGGAUUACCCAAAUUGCCCAACUGCCAGAGCUCCCAGGAGUUGACCUGUGGCGAGGGA ........((.(((((((.(((....((((((((..((.....((((....)))).(((((((((.....))...((....)).)))))))))..))))))))))))))))))..))... (-37.60 = -37.56 + -0.04)

| Location | 3,362,986 – 3,363,106 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.83 |
| Mean single sequence MFE | -35.83 |
| Consensus MFE | -27.16 |
| Energy contribution | -27.36 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
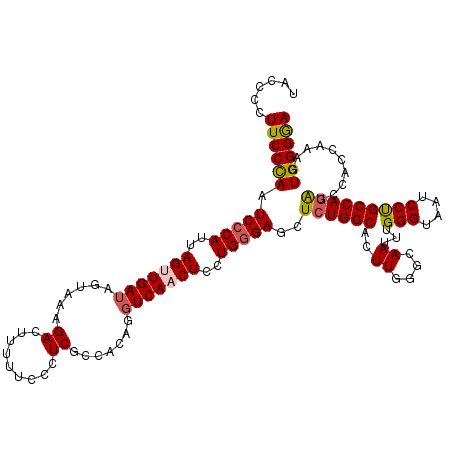
>3L_DroMel_CAF1 3362986 120 - 23771897 UACCCCUUCCCAAUCCCAUCAGCUGAUGGUAAAGACUUUUUCCCUCGCCACAGGUCAACUCCUGAGAGCUCUGGCAUUUGUCCAAUUUGGGUAAUCCUGCCAGAUCCACCAAAGUGGGGA ..((((.........(((((....))))).....(((((.......(((.((((......)))).).))(((((((.((..((.....))..))...)))))))......))))))))). ( -38.50) >DroSec_CAF1 30144 120 - 1 UACCCCUUCCCAAUCCCAUUAGUUGAUAGUAAAGACUUUUUCCCUCGCCACAGGUCAACUCCUGGGAGCUCUGGCACUUGGGCAAUUUGGGUAAUCCUGCCAGAUCCACCAAUGUGGGGA ..((((..((((((((((..(((((((......((.........)).......)))))))..)))))((....))..)))))....((((...(((......)))...))))...)))). ( -34.42) >DroSim_CAF1 27845 120 - 1 UACCCCUUCCCAAUCCCAUCAGCUGAUAAUAAAGACUUUUUCCCUCGCCACAGGUCAACUCCUGGGAGCUCUGGCACUUGGGCAAUUUGGGUAAUCCUGCCAUAUCCACCAAUGUGGGGA (((((.(((((((..(((..((((((.((........)).)).....((.((((......)))))))))).)))...))))).))...))))).(((..((((((......))))))))) ( -30.90) >DroEre_CAF1 28942 120 - 1 UACCCCUUCCUAGUCCCAUUAGUUGAUAGUAAAGACUCUUUCCCUCGCCACAGGUCAACUCCUGGGAGCUCUGGCAGUUGGGCAACUUGGGUAAUCCCGCCAGGUCCACCAAAGUGGGAA (((((........(((((..(((((((......((.........)).......)))))))..)))))((((........)))).....))))).((((((..((....))...)))))). ( -37.62) >DroYak_CAF1 28727 120 - 1 UACCACUUCCCAGUCCCAUUAGUUGAGGAUAAAGACUUUUUCCCUCGCCACAGGUCAACUCCUGGGAGCUCUGGCAGUUGGGCAAUUUGGGUAAUCCCGCCAGAUCCACCAAGGUGGGAA ........(((((.((((......((((..(((....)))..))))(((.((((......))))((....)))))...))))....)))))...(((((((...........))))))). ( -37.70) >consensus UACCCCUUCCCAAUCCCAUUAGUUGAUAGUAAAGACUUUUUCCCUCGCCACAGGUCAACUCCUGGGAGCUCUGGCACUUGGGCAAUUUGGGUAAUCCUGCCAGAUCCACCAAAGUGGGGA ......((((((.(((((..(((((((......((.........)).......)))))))..)))))..((((((..((....))...(((....)))))))))..........)))))) (-27.16 = -27.36 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:59 2006