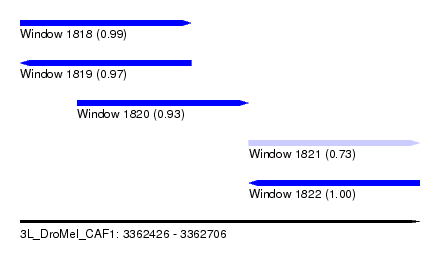
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,362,426 – 3,362,706 |
| Length | 280 |
| Max. P | 0.997614 |

| Location | 3,362,426 – 3,362,546 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.83 |
| Mean single sequence MFE | -47.38 |
| Consensus MFE | -43.52 |
| Energy contribution | -43.44 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3362426 120 + 23771897 GCUGCCGGACAUCGAGUCCGUGGCGGAGGCAUUGCGCUUUGUGGGCUCACGGAGCAGGCACAGCCAGGAACCAUUUUUUCUGGCCAUGGGCUUCCACAAGCCGCACAUUAAUUUCCGGUU ...((((((.((..(....(((((((((((.....((((((((....))))))))...((..((((((((......))))))))..)).))))))....)))))...)..)).)))))). ( -49.10) >DroSec_CAF1 29584 120 + 1 GCUGCCGGACAUUGAGUCCGUGGCGGAGGCACUGCGCUUUGUGGGCUCACGGAGCAGGCACAGCCAGGAACCAUUUUUCCUGGCCAUGGGUUUCCACAAGCCGCACAUUAAUUUCCGGUU ...((((((.(((((....(((((((((((.....((((((((....))))))))...((..((((((((......))))))))..)).))))))....)))))...))))).)))))). ( -52.20) >DroSim_CAF1 27285 120 + 1 GCUGCCGGACAUAGAGUCCGUGGCGGAGGCACUGCGCUUUGUGGGCUCACGGAGCAGGCACAGCCAGGAACCAUUUUUUCUGGCCAUGGGCUUCCACAAGCCGCACAUUAAUUUCCGGUU ...(((((((.....))))(((((((((((.....((((((((....))))))))...((..((((((((......))))))))..)).))))))....)))))............))). ( -48.60) >DroEre_CAF1 28382 120 + 1 GCUGCCGGACAUCGAGUCCGUGGCGGAGGCACUGCGCUUCGUGGAGUCACGGAGCACGCACAACCAGGAACCAUUUUUUCUGGCCAUGGGUUUCCACAAGCCACACAUUAAUUUCCGAUU ((..(.((((.....)))))..))((((((..(((((((((((....))))))))..)))....((((((......))))))))).(((((((....))))).))........))).... ( -42.60) >DroYak_CAF1 28167 120 + 1 ACUGCCGGACAUCGAGUCCGUGGCGGAGGCACUGCGCUUUGUGGAGUCACGGAGCACGCGGAAGCAGGAACCCUUUUUCCUGGCCAUGGGUUUUCACAAGCCUCACAUUAAUUUCCGGUU ...((((((.((..(.((((((((((.....))))((((((((....)))))))).))))))..((((((......)))))).....((((((....))))))....)..)).)))))). ( -44.40) >consensus GCUGCCGGACAUCGAGUCCGUGGCGGAGGCACUGCGCUUUGUGGGCUCACGGAGCAGGCACAGCCAGGAACCAUUUUUUCUGGCCAUGGGUUUCCACAAGCCGCACAUUAAUUUCCGGUU ...(((((((.....))))(((((((((((..(((((((((((....))))))))..)))..((((((((......)))))))).....))))))....)))))............))). (-43.52 = -43.44 + -0.08)

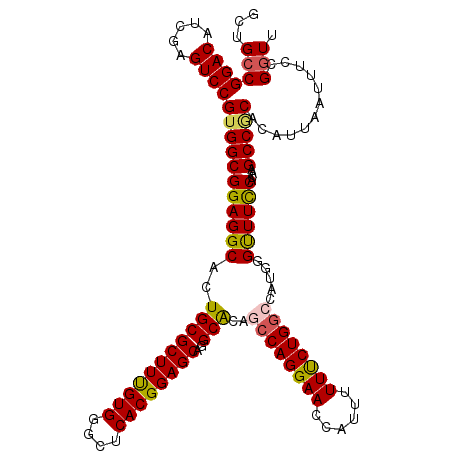
| Location | 3,362,426 – 3,362,546 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.83 |
| Mean single sequence MFE | -46.39 |
| Consensus MFE | -38.90 |
| Energy contribution | -39.46 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3362426 120 - 23771897 AACCGGAAAUUAAUGUGCGGCUUGUGGAAGCCCAUGGCCAGAAAAAAUGGUUCCUGGCUGUGCCUGCUCCGUGAGCCCACAAAGCGCAAUGCCUCCGCCACGGACUCGAUGUCCGGCAGC ....(((......((((.((((..((((.((.(((((((((............)))))))))...))))))..))))))))..((.....)).)))(((..((((.....)))))))... ( -48.50) >DroSec_CAF1 29584 120 - 1 AACCGGAAAUUAAUGUGCGGCUUGUGGAAACCCAUGGCCAGGAAAAAUGGUUCCUGGCUGUGCCUGCUCCGUGAGCCCACAAAGCGCAGUGCCUCCGCCACGGACUCAAUGUCCGGCAGC ....(((......((((.((((..((((....((((((((((((......)))))))))))).....))))..))))))))..((.....)).)))(((..((((.....)))))))... ( -52.60) >DroSim_CAF1 27285 120 - 1 AACCGGAAAUUAAUGUGCGGCUUGUGGAAGCCCAUGGCCAGAAAAAAUGGUUCCUGGCUGUGCCUGCUCCGUGAGCCCACAAAGCGCAGUGCCUCCGCCACGGACUCUAUGUCCGGCAGC ....(((......((((.((((..((((.((.(((((((((............)))))))))...))))))..))))))))..((.....)).)))(((..((((.....)))))))... ( -48.50) >DroEre_CAF1 28382 120 - 1 AAUCGGAAAUUAAUGUGUGGCUUGUGGAAACCCAUGGCCAGAAAAAAUGGUUCCUGGUUGUGCGUGCUCCGUGACUCCACGAAGCGCAGUGCCUCCGCCACGGACUCGAUGUCCGGCAGC ....((((.......(.(((((.((((....))))))))).)........)))).((..(..((((((.((((....)))).))))).)..)..))(((..((((.....)))))))... ( -44.76) >DroYak_CAF1 28167 120 - 1 AACCGGAAAUUAAUGUGAGGCUUGUGAAAACCCAUGGCCAGGAAAAAGGGUUCCUGCUUCCGCGUGCUCCGUGACUCCACAAAGCGCAGUGCCUCCGCCACGGACUCGAUGUCCGGCAGU ..(((((........((((....((....))((.((((((((((......))))))....((((((((..(((....)))..))))).))).....)))).)).))))...))))).... ( -37.60) >consensus AACCGGAAAUUAAUGUGCGGCUUGUGGAAACCCAUGGCCAGAAAAAAUGGUUCCUGGCUGUGCCUGCUCCGUGAGCCCACAAAGCGCAGUGCCUCCGCCACGGACUCGAUGUCCGGCAGC ....(((......((((.((((..((((....(((((((((............))))))))).....))))..))))))))..((.....)).)))(((..((((.....)))))))... (-38.90 = -39.46 + 0.56)

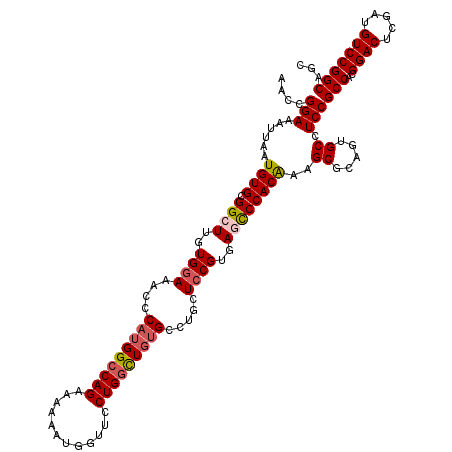
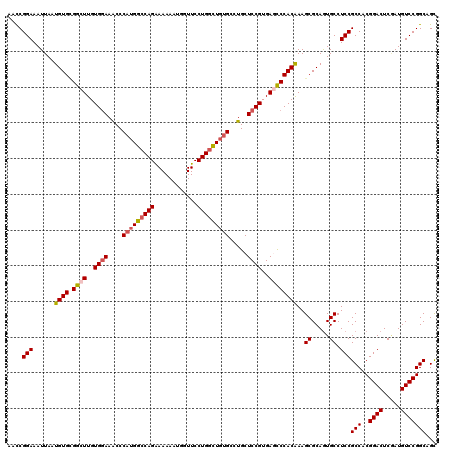
| Location | 3,362,466 – 3,362,586 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.50 |
| Mean single sequence MFE | -34.94 |
| Consensus MFE | -27.88 |
| Energy contribution | -27.80 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3362466 120 + 23771897 GUGGGCUCACGGAGCAGGCACAGCCAGGAACCAUUUUUUCUGGCCAUGGGCUUCCACAAGCCGCACAUUAAUUUCCGGUUCCCACGGCAAUUCCUUUCGAGAUUCAACCUUUCCCAGUUU (((((..(.(((((....((..((((((((......))))))))..))(((((....)))))..........))))))..)))))((.(((..(....)..)))...))........... ( -33.50) >DroSec_CAF1 29624 120 + 1 GUGGGCUCACGGAGCAGGCACAGCCAGGAACCAUUUUUCCUGGCCAUGGGUUUCCACAAGCCGCACAUUAAUUUCCGGUUCCCACUGCAAUUCCUAUCGAGAUUCCACCUUUCCCAGUUU ((((.(((..(((((((.....((((((((......))))))))..((((........(((((............)))))))))))))...)))....)))...))))............ ( -35.00) >DroSim_CAF1 27325 120 + 1 GUGGGCUCACGGAGCAGGCACAGCCAGGAACCAUUUUUUCUGGCCAUGGGCUUCCACAAGCCGCACAUUAAUUUCCGGUUCCCACGGCAAUUUCUAUCGAGAUUUCACCUUUCCCAGUUU (((((..(.(((((....((..((((((((......))))))))..))(((((....)))))..........))))))..)))))((.((((((....))))))...))........... ( -36.80) >DroEre_CAF1 28422 120 + 1 GUGGAGUCACGGAGCACGCACAACCAGGAACCAUUUUUUCUGGCCAUGGGUUUCCACAAGCCACACAUUAAUUUCCGAUUCCCACGGCAAUUUCUAUCAAGAUUCCACCUUUCCGACUUU ((((((((..((((.((.((...(((((((......)))))))...)).))))))....(((.......................)))............))))))))............ ( -30.30) >DroYak_CAF1 28207 120 + 1 GUGGAGUCACGGAGCACGCGGAAGCAGGAACCCUUUUUCCUGGCCAUGGGUUUUCACAAGCCUCACAUUAAUUUCCGGUUUCCACGGCAAUUUCUGUCAAGAUUCCACCUCUCCCAGUUU ((((((((..((((.((.((((((((((((......)))))).....((((((....))))))........))))))))))))..((((.....))))..))))))))............ ( -39.10) >consensus GUGGGCUCACGGAGCAGGCACAGCCAGGAACCAUUUUUUCUGGCCAUGGGUUUCCACAAGCCGCACAUUAAUUUCCGGUUCCCACGGCAAUUUCUAUCGAGAUUCCACCUUUCCCAGUUU (((((....(((((....((..((((((((......))))))))..))(((((....)))))..........)))))...)))))................................... (-27.88 = -27.80 + -0.08)

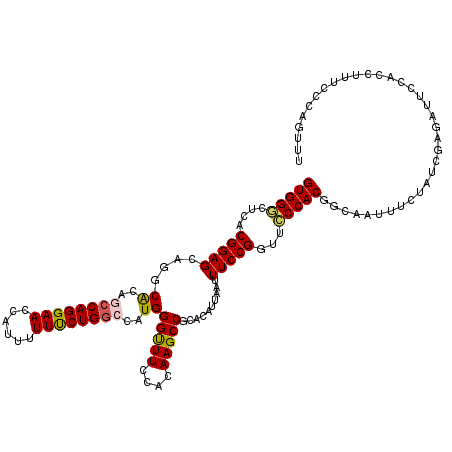
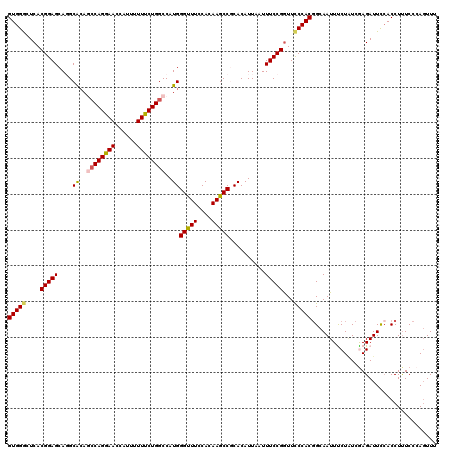
| Location | 3,362,586 – 3,362,706 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.58 |
| Mean single sequence MFE | -40.84 |
| Consensus MFE | -37.50 |
| Energy contribution | -38.10 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.731650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3362586 120 + 23771897 UACAAUUACACGGAGGAUAGCCUGAAGCCGCCGGAUAUGCCGGCAGUGGCCUGGAAUCCUUACACAGAUGUGCGGGCCAGGGAUGACUUCAAGCACUCCAACAUAUCCUUUCCCUACGGA ...........((((....((.((((((((((((.....)))))..(((((((.((((........))).).))))))).))....))))).)).))))...........(((....))) ( -38.10) >DroSec_CAF1 29744 120 + 1 UACAACUACACGGAGGACAGCCUGAAGCCGCCGGACAUGCCGGCAGUGGCCUGGAAUCCCUACACGGAUGUGCGGGCCAGGGAUGACUUCAAGCACUCCAACAUAUCCUUUCCCUACGGA ...........((((....((.((((((((((((.....)))))..(((((((.(((((......)))).).))))))).))....))))).)).))))...........(((....))) ( -41.50) >DroSim_CAF1 27445 120 + 1 UACAACUACACGGAGGACAGCCUGAAGCCGCCGGACAUGCCGGCAGUGGCCUGGAAUCCCUACACGGAUGUGCGGGCCAGAGAUGACUUCAAGCACUCCAACAUAUCCUUUCCCUACGGA ...........((((....((.(((((.((((((.....)))))..(((((((.(((((......)))).).))))))).....).))))).)).))))...........(((....))) ( -41.40) >DroEre_CAF1 28542 120 + 1 UACAACUACACGGAGGACAGCCUGAAGCCGCCGGAUAUGCCGGCAGUAGCCUGGAAUCCGUACACGGAUGUGCGGGCCAGGGAUGACUUCAAGCACUCCAAUAUAUCGUUUCCCUACGGA ...........((((....((.(((((.((((((.....))))).....(((((...(((((((....))))))).)))))...).))))).)).))))......((((......)))). ( -45.00) >DroYak_CAF1 28327 120 + 1 UACAACUACACGGAGGACAGCCUGAAGCCGCCGGAUAUGCCGGCAGUGGCCUGGAAUCCCUACACGGAUGUGCGGGCCAGGGAUGACUUCAAGCACACCAAUAUAUCGUUUCCCUACGGC ..............((...((.((((((((((((.....)))))..(((((((.(((((......)))).).))))))).))....))))).))...))......((((......)))). ( -38.20) >consensus UACAACUACACGGAGGACAGCCUGAAGCCGCCGGAUAUGCCGGCAGUGGCCUGGAAUCCCUACACGGAUGUGCGGGCCAGGGAUGACUUCAAGCACUCCAACAUAUCCUUUCCCUACGGA ...........((((....((.(((((.((((((.....)))))..(((((((.(((((......)))).).))))))).....).))))).)).))))..................... (-37.50 = -38.10 + 0.60)

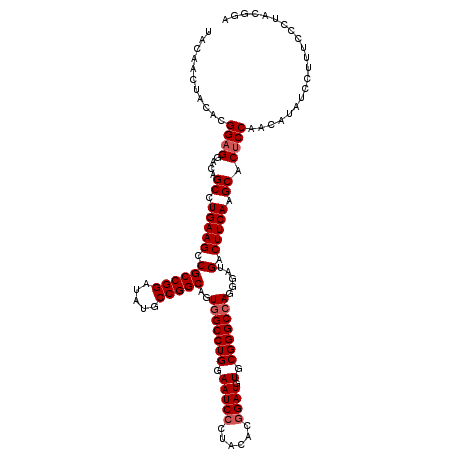
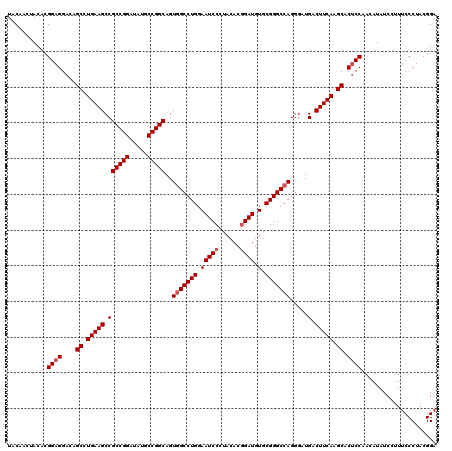
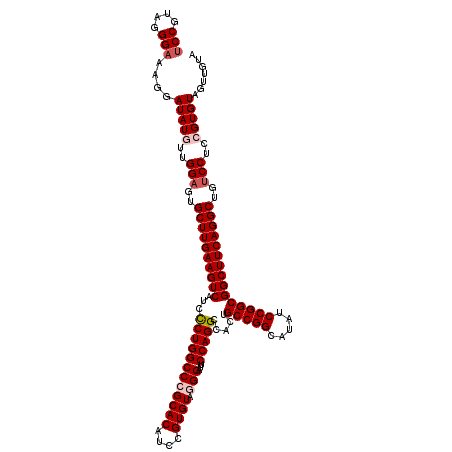
| Location | 3,362,586 – 3,362,706 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.58 |
| Mean single sequence MFE | -48.88 |
| Consensus MFE | -44.32 |
| Energy contribution | -45.36 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.997614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3362586 120 - 23771897 UCCGUAGGGAAAGGAUAUGUUGGAGUGCUUGAAGUCAUCCCUGGCCCGCACAUCUGUGUAAGGAUUCCAGGCCACUGCCGGCAUAUCCGGCGGCUUCAGGCUAUCCUCCGUGUAAUUGUA (((....)))((..(((((..(((..((((((((((...(((((.((((((....))))..))...))))).....(((((.....)))))))))))))))..)))..)))))..))... ( -47.70) >DroSec_CAF1 29744 120 - 1 UCCGUAGGGAAAGGAUAUGUUGGAGUGCUUGAAGUCAUCCCUGGCCCGCACAUCCGUGUAGGGAUUCCAGGCCACUGCCGGCAUGUCCGGCGGCUUCAGGCUGUCCUCCGUGUAGUUGUA (((....)))....(((((..(((..((((((((((...((((((((((((....)))).)))...))))).....(((((.....)))))))))))))))..)))..)))))....... ( -51.10) >DroSim_CAF1 27445 120 - 1 UCCGUAGGGAAAGGAUAUGUUGGAGUGCUUGAAGUCAUCUCUGGCCCGCACAUCCGUGUAGGGAUUCCAGGCCACUGCCGGCAUGUCCGGCGGCUUCAGGCUGUCCUCCGUGUAGUUGUA (((....)))....(((((..(((..((((((((((...((((((((((((....)))).)))...))))).....(((((.....)))))))))))))))..)))..)))))....... ( -48.70) >DroEre_CAF1 28542 120 - 1 UCCGUAGGGAAACGAUAUAUUGGAGUGCUUGAAGUCAUCCCUGGCCCGCACAUCCGUGUACGGAUUCCAGGCUACUGCCGGCAUAUCCGGCGGCUUCAGGCUGUCCUCCGUGUAGUUGUA (((....))).(((((((((.((((.((((((((((...(((((.(((.(((....))).)))...))))).....(((((.....)))))))))))))))....)))))))).))))). ( -49.30) >DroYak_CAF1 28327 120 - 1 GCCGUAGGGAAACGAUAUAUUGGUGUGCUUGAAGUCAUCCCUGGCCCGCACAUCCGUGUAGGGAUUCCAGGCCACUGCCGGCAUAUCCGGCGGCUUCAGGCUGUCCUCCGUGUAGUUGUA .((....))..(((((((((.((...((((((((((...((((((((((((....)))).)))...))))).....(((((.....)))))))))))))))...))...)))).))))). ( -47.60) >consensus UCCGUAGGGAAAGGAUAUGUUGGAGUGCUUGAAGUCAUCCCUGGCCCGCACAUCCGUGUAGGGAUUCCAGGCCACUGCCGGCAUAUCCGGCGGCUUCAGGCUGUCCUCCGUGUAGUUGUA (((....)))....(((((..(((..((((((((((...((((((((((((....)))).)))...))))).....(((((.....)))))))))))))))..)))..)))))....... (-44.32 = -45.36 + 1.04)

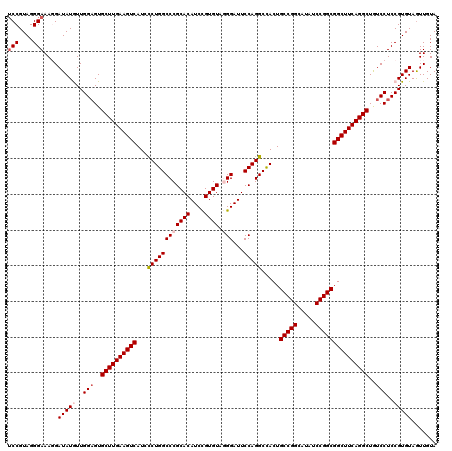
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:57 2006