| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,358,774 – 3,358,894 |
| Length | 120 |
| Max. P | 0.821915 |

| Location | 3,358,774 – 3,358,894 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.33 |
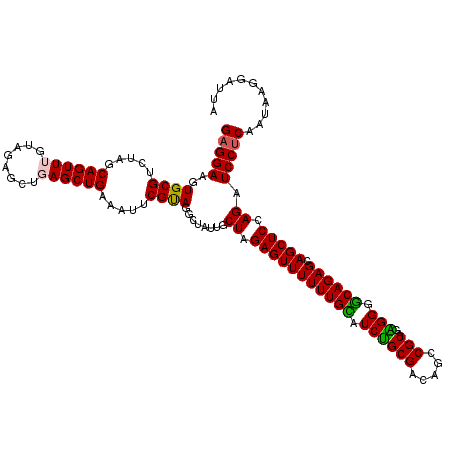
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -24.14 |
| Energy contribution | -24.62 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.515817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3358774 120 + 23771897 UAAUCCUCAUUGAGGAUCUGGAGCUGCUCUGCCCCUCAACGGCUGUCGCAGAUGCAAAAAACUCUAGCAAUAGCCUACGAAUUUCAGCUGAGCUCUACAAACUGCUAGACGCACUUCCUC ...........(((((..(((((((...(((....((...((((((.(((((..........))).)).))))))...))....)))...))))))).....(((.....)))..))))) ( -29.50) >DroSec_CAF1 25958 120 + 1 UAAUCCUCAUUGAGGAUCUGGAGCUGCUCUGCCCCUCUACGGCUGUCGCAGAUGCAAAAAACUCUAGCAAUAGCCUACGAAUUUCAGCUCAGCUCUACAAACUGCUGGACGCACUUCCUC ...........(((((..((((((((..(((....((...((((((.(((((..........))).)).))))))...))....)))..)))))))).....(((.....)))..))))) ( -33.20) >DroSim_CAF1 23647 120 + 1 UAAUCCUUAUUGAGGAUCUGGAGCUGCUCUGCCCCUCUACGGCUGUCGCAGAUGCAAAAAACUCUAGCAAUAGCCUACGAAUUUCAGCUGAGCUCUACAAACUGCUAGACGCACUUCCUC ...........(((((..(((((((...(((....((...((((((.(((((..........))).)).))))))...))....)))...))))))).....(((.....)))..))))) ( -29.50) >DroEre_CAF1 24523 120 + 1 UGAUACUUAUUGAGGAUCUGGAGCUGCUCUGCCCCCUAACGGGUGUCGCAGAUACAAAAAACUCAAGCAAUUCUCUACGAAUUUCAGCUCAGCUAUACAAACUGAUAGACGCACUUCCUC ...........(((((..((((((((.(((((((((....))).)..)))))................(((((.....))))).)))))).(((((........)))).).))..))))) ( -30.80) >DroYak_CAF1 24511 120 + 1 UGAUACUUAUUGAGGAUCUGGAGCUGCUCUGUCCCCUUACGGGUGUCGCGGACACAAAAAACUCUAGCAAUUCUCUGCGGAUAUCAGCUCAGCUCUACAAACUGCUAGACGCACUUCCGC ...........((((((((((((.....((((......))))(((((...)))))......))))))..)))))).(((((.....(((((((..........))).)).))...))))) ( -30.50) >consensus UAAUCCUUAUUGAGGAUCUGGAGCUGCUCUGCCCCUCUACGGCUGUCGCAGAUGCAAAAAACUCUAGCAAUAGCCUACGAAUUUCAGCUCAGCUCUACAAACUGCUAGACGCACUUCCUC ...........(((((..((((((((..(((....((...((((((.(((((..........))).)).))))))...))....)))..)))))))).....(((.....)))..))))) (-24.14 = -24.62 + 0.48)

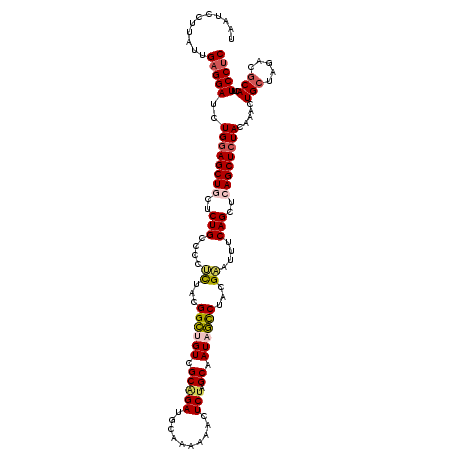
| Location | 3,358,774 – 3,358,894 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.33 |
| Mean single sequence MFE | -40.12 |
| Consensus MFE | -31.00 |
| Energy contribution | -30.64 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3358774 120 - 23771897 GAGGAAGUGCGUCUAGCAGUUUGUAGAGCUCAGCUGAAAUUCGUAGGCUAUUGCUAGAGUUUUUUGCAUCUGCGACAGCCGUUGAGGGGCAGAGCAGCUCCAGAUCCUCAAUGAGGAUUA .(.(((.(((.....))).))).).(((((...(((...((((..((((.((((.(((((.....)).))))))).))))..))))...)))...)))))..(((((((...))))))). ( -40.80) >DroSec_CAF1 25958 120 - 1 GAGGAAGUGCGUCCAGCAGUUUGUAGAGCUGAGCUGAAAUUCGUAGGCUAUUGCUAGAGUUUUUUGCAUCUGCGACAGCCGUAGAGGGGCAGAGCAGCUCCAGAUCCUCAAUGAGGAUUA .(.(((.(((.....))).))).).((((((..(((...(((...((((.((((.(((((.....)).))))))).))))...)))...)))..))))))..(((((((...))))))). ( -46.40) >DroSim_CAF1 23647 120 - 1 GAGGAAGUGCGUCUAGCAGUUUGUAGAGCUCAGCUGAAAUUCGUAGGCUAUUGCUAGAGUUUUUUGCAUCUGCGACAGCCGUAGAGGGGCAGAGCAGCUCCAGAUCCUCAAUAAGGAUUA ..(((..(((.((((((((((((..((..((....))..))..)))))...)))))))....(((((.((((((.....))))))...))))))))..))).((((((.....)))))). ( -40.20) >DroEre_CAF1 24523 120 - 1 GAGGAAGUGCGUCUAUCAGUUUGUAUAGCUGAGCUGAAAUUCGUAGAGAAUUGCUUGAGUUUUUUGUAUCUGCGACACCCGUUAGGGGGCAGAGCAGCUCCAGAUCCUCAAUAAGUAUCA (((((..((((....(((((((........)))))))....))))........((.((((....(((.(((((....(((....))).)))))))))))).)).)))))........... ( -35.20) >DroYak_CAF1 24511 120 - 1 GCGGAAGUGCGUCUAGCAGUUUGUAGAGCUGAGCUGAUAUCCGCAGAGAAUUGCUAGAGUUUUUUGUGUCCGCGACACCCGUAAGGGGACAGAGCAGCUCCAGAUCCUCAAUAAGUAUCA (((((..(((.....))).))))).((((((..(((.....(((((((((((.....))))))))))).........(((....)))..)))..))))))..(((.((.....)).))). ( -38.00) >consensus GAGGAAGUGCGUCUAGCAGUUUGUAGAGCUGAGCUGAAAUUCGUAGGCUAUUGCUAGAGUUUUUUGCAUCUGCGACAGCCGUAGAGGGGCAGAGCAGCUCCAGAUCCUCAAUAAGGAUUA (((((..((((.....((((((........)))))).....))))........((.(((((((((((.((((((.....)))..))).)))))).))))).)).)))))........... (-31.00 = -30.64 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:48 2006