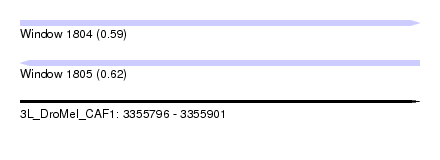
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,355,796 – 3,355,901 |
| Length | 105 |
| Max. P | 0.615336 |

| Location | 3,355,796 – 3,355,901 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.88 |
| Mean single sequence MFE | -47.08 |
| Consensus MFE | -37.33 |
| Energy contribution | -37.28 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3355796 105 + 23771897 CCGCCGGGUGGCGCUCCUGCUGCACC------UCCCAUCUGGCGGUACGGGGAACCGCUUCUUUUAAAGCCACCGCCCGGCGGAAGGCCCAUGCCA---------UGAUGGUGACGGUAC ((((((((.((.((.......)).))------.)))....((((((..((....))((((......)))).)))))).)))))...(((...(((.---------....)))...))).. ( -45.90) >DroSec_CAF1 22977 105 + 1 CCGCCGGGUGGCGCUCCCGCUGCACC------UCCCAUCUGGCGGUACGGAGAACCGCUUCUCCUAAAGCCACCGCCCGGCGGAAGGCCCAUGCCA---------UGAUGGUGACGGUAC (((((((((((.((.......))...------.......((((..((.((((((....))))))))..)))))))))))))))...(((...(((.---------....)))...))).. ( -47.60) >DroSim_CAF1 20665 105 + 1 CCGCCGGGUGGCGCUCCCGCUGCACC------UCCCAUCUGGCGGUACGGAGAACCGCUUCUCUUAAAGCCACCGCCCGGCGGAAGGCCCAUGCCA---------UGAUGGUGACGGUAC (((((((((((.((.......))...------.......((((..((.((((((....))))))))..)))))))))))))))...(((...(((.---------....)))...))).. ( -44.90) >DroEre_CAF1 21556 105 + 1 CCGCCGGGUGGUGCUCCUGAUGCACC------UCCCAUCUGGCGGUAGGGAGAACCGCUUCUCUUAAAGCCACCGCCCGGCGGAAGGCCCAUGCCA---------UGAUGGUGACGGUAC ((((((((((((((((((.....(((------.((.....)).))).)))))....((((......)))))))))))))))))...(((...(((.---------....)))...))).. ( -49.00) >DroYak_CAF1 21551 105 + 1 CCGCCGGGUGGUGCUCCUGCUGCACC------UCCCAUCUGGCGGUAGGGAGAACCGCUUCUCUUAAAGCCACCGCCCGGCGGAAGGCCCAUGCCA---------UGAUGGUGACGAUAC (((((((((((((((((((((((...------.........))))))))(((((....)))))....)).)))))))))))))..(((....))).---------...((....)).... ( -52.10) >DroAna_CAF1 23691 117 + 1 CCACCUGGUG---CUCCGCCAGCACCACCAGCACCCAUCUGGCGAUAGGGUGAGCCACUACGCUUGAAGCCACCACCUGGCGGUAUGCCCAUACCACCAUGAUGAUGAUGGUGGCGAUAC .....(((((---((.....)))))))....((((((((....))).))))).(((((((.((.....))((.(((.(((.(((((....))))).))).).)).)).)))))))..... ( -43.00) >consensus CCGCCGGGUGGCGCUCCUGCUGCACC______UCCCAUCUGGCGGUACGGAGAACCGCUUCUCUUAAAGCCACCGCCCGGCGGAAGGCCCAUGCCA_________UGAUGGUGACGGUAC (((((((((((.((.......))................((((..((.((((((....))))))))..)))))))))))))))...(((...((((............))))...))).. (-37.33 = -37.28 + -0.05)

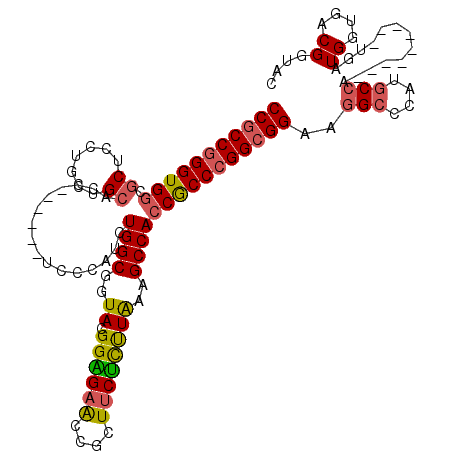
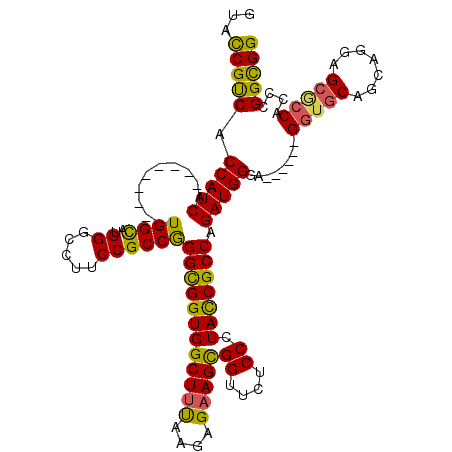
| Location | 3,355,796 – 3,355,901 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.88 |
| Mean single sequence MFE | -49.37 |
| Consensus MFE | -43.38 |
| Energy contribution | -42.55 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3355796 105 - 23771897 GUACCGUCACCAUCA---------UGGCAUGGGCCUUCCGCCGGGCGGUGGCUUUAAAAGAAGCGGUUCCCCGUACCGCCAGAUGGGA------GGUGCAGCAGGAGCGCCACCCGGCGG (..((((..((....---------.)).))))..)..((((((((.(((.(((((....))))).(((((..(((((.((.....)).------)))))....)))))))).)))))))) ( -48.20) >DroSec_CAF1 22977 105 - 1 GUACCGUCACCAUCA---------UGGCAUGGGCCUUCCGCCGGGCGGUGGCUUUAGGAGAAGCGGUUCUCCGUACCGCCAGAUGGGA------GGUGCAGCGGGAGCGCCACCCGGCGG (((((.((.(((((.---------((((..(((((..((((...)))).)))))..((((((....)))))).....)))))))))))------))))).((....))(((....))).. ( -51.20) >DroSim_CAF1 20665 105 - 1 GUACCGUCACCAUCA---------UGGCAUGGGCCUUCCGCCGGGCGGUGGCUUUAAGAGAAGCGGUUCUCCGUACCGCCAGAUGGGA------GGUGCAGCGGGAGCGCCACCCGGCGG (..((((..((....---------.)).))))..)..((((((((.(((.(((((....))))).((((((((((((.((.....)).------))))).).))))))))).)))))))) ( -49.60) >DroEre_CAF1 21556 105 - 1 GUACCGUCACCAUCA---------UGGCAUGGGCCUUCCGCCGGGCGGUGGCUUUAAGAGAAGCGGUUCUCCCUACCGCCAGAUGGGA------GGUGCAUCAGGAGCACCACCCGGCGG (..((((..((....---------.)).))))..)..(((((.((((((((((((....)))))((....)).)))))))....(((.------(((((.......))))).)))))))) ( -46.70) >DroYak_CAF1 21551 105 - 1 GUAUCGUCACCAUCA---------UGGCAUGGGCCUUCCGCCGGGCGGUGGCUUUAAGAGAAGCGGUUCUCCCUACCGCCAGAUGGGA------GGUGCAGCAGGAGCACCACCCGGCGG ...............---------.(((....)))..(((((.((((((((((((....)))))((....)).)))))))....(((.------(((((.......))))).)))))))) ( -45.80) >DroAna_CAF1 23691 117 - 1 GUAUCGCCACCAUCAUCAUCAUGGUGGUAUGGGCAUACCGCCAGGUGGUGGCUUCAAGCGUAGUGGCUCACCCUAUCGCCAGAUGGGUGCUGGUGGUGCUGGCGGAG---CACCAGGUGG .....(((((...(((((((.(((((((((....))))))))))))))))((.....))...))))).(((((((.((((.....)))).)))(((((((.....))---))))))))). ( -54.70) >consensus GUACCGUCACCAUCA_________UGGCAUGGGCCUUCCGCCGGGCGGUGGCUUUAAGAGAAGCGGUUCUCCCUACCGCCAGAUGGGA______GGUGCAGCAGGAGCGCCACCCGGCGG ...(((((.(((((..........((((..((.....))))))((((((((((((....)))))((....)).))))))).)))))........(((((.......)))))....))))) (-43.38 = -42.55 + -0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:39 2006