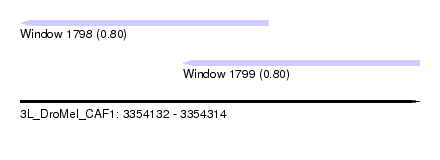
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,354,132 – 3,354,314 |
| Length | 182 |
| Max. P | 0.800457 |

| Location | 3,354,132 – 3,354,245 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.33 |
| Mean single sequence MFE | -20.57 |
| Consensus MFE | -15.85 |
| Energy contribution | -15.58 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.800457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3354132 113 - 23771897 GUGUUUGGCCCACCAGUUGGGCACGCGUCAAUCCC-CCCAUCGAUUCCACUCAGUGACUGCCACAUUUUCACGCUUCACCAAAGCUCAGACCGUCGACAAACACGA-----AA-AUACAU ((((((((((((.....)))))(((.(((......-.................((((...........))))((((.....))))...))))))...)))))))..-----..-...... ( -28.30) >DroSec_CAF1 21318 113 - 1 CUGUUUGCCCCACCAGUUGGGCUCCUGUCAAUCCC-CCCAUCGAUUCCACUCAAUGACUGCCACAUUUUCACGCUUCACCAAAGCUCAGACCGUCGACAAACACGC-----AA-AUACAU .(((((((((((.....))))....((((......-..(((.((......)).)))................((((.....))))..........)))).....))-----))-)))... ( -18.10) >DroSim_CAF1 19001 113 - 1 CUGUUUGGCCCACCAGUUGGGCACCUGUCAAUCCC-CCCAUCGAUUCCACUCAAUGACUGCCACAUUUUCACGCUUCACCAAAGCUCAGACCGUCGACAAACACGC-----AA-AUACAU .(((((((((((.....)))))....(((......-..(((.((......)).)))................((((.....))))...)))......))))))...-----..-...... ( -19.90) >DroEre_CAF1 19883 109 - 1 CUGUUUGGCCCACUAGUUGGGCAC----CAAUCCC-CCCAUCGAUUUCACUCAAUGACUGCCACAUUUUCACGCUUCACCAGAGCUCAGACCGUCGACAAACACUC-----AA-UUACAU .(((((((((((.....)))))..----.......-....(((((.((....((((.......)))).....((((.....))))...))..)))))))))))...-----..-...... ( -20.90) >DroYak_CAF1 19878 111 - 1 CUGUUUGGCCCACUAGCUGGGCAC----CAAUCCCCCCCAUCGAUUUCACUCAAUGACUGCCACAUUUUCACGCUUCACCAGAGCUCAGACCGUCGACAAACACUC-----AAAAUACAU .(((((((((((.....)))))..----............(((((.((....((((.......)))).....((((.....))))...))..)))))))))))...-----......... ( -20.90) >DroAna_CAF1 22110 108 - 1 CUGUUUCGACUCCUUCAAAGGUCC----AUAUAUC-ACCAUCGAUUCCAUUCAAAGACUGCCACUCUUUCGAACUCCCACGGAGU------CCUCGACCAACAUCCGUAGUAA-UGACAU .......((((........)))).----.......-..............(((...(((((.......(((((((((...)))))------..)))).........)))))..-)))... ( -15.29) >consensus CUGUUUGGCCCACCAGUUGGGCAC____CAAUCCC_CCCAUCGAUUCCACUCAAUGACUGCCACAUUUUCACGCUUCACCAAAGCUCAGACCGUCGACAAACACGC_____AA_AUACAU .(((((((((((.....)))))..................(((((.......((((.......)))).....((((.....)))).......)))))))))))................. (-15.85 = -15.58 + -0.27)
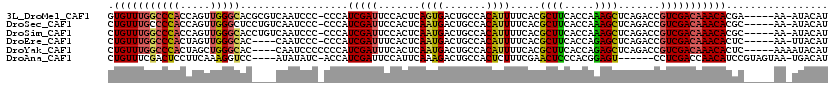
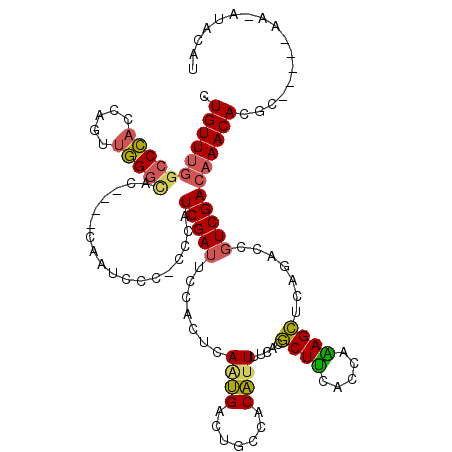
| Location | 3,354,206 – 3,354,314 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.22 |
| Mean single sequence MFE | -32.43 |
| Consensus MFE | -21.71 |
| Energy contribution | -23.02 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3354206 108 - 23771897 ----AUUUUGGACCGGAGUGAUGCGCUCCGGGUCCCU-UUGGCUCAAUAGCAAUA------CGAACCGUAGCAAUCCGAAGUGUUUGGCCCACCAGUUGGGCACGCGUCAAUCCC-CCCA ----....(((...(((.(((((((((.(((.((...-...(((....)))....------.)).))).)))........(((((..((......))..))))))))))).))).-.))) ( -36.40) >DroSec_CAF1 21392 108 - 1 ----AUUUUGUCCCGGAGUGAUGCGCUCCGGGUCCCU-UUGGCUCAAUAGCAAUA------CGAACCGUAGCAAUCCGAACUGUUUGCCCCACCAGUUGGGCUCCUGUCAAUCCC-CCCA ----.....(.(((((((((...))))))))).)...-(((((......((..((------((...))))))...((.(((((..........))))).)).....)))))....-.... ( -29.20) >DroSim_CAF1 19075 108 - 1 ----AUUUUGGCCCGGAGUGAUGCGCUCCGGGUCCCU-UUGGCUCAAUAGCAAUA------CGAACCGUAGCAAUCCGAACUGUUUGGCCCACCAGUUGGGCACCUGUCAAUCCC-CCCA ----.....(((((((((((...)))))))))))...-(((((............------(((((.((..........)).)))))(((((.....)))))....)))))....-.... ( -35.50) >DroEre_CAF1 19957 105 - 1 ----AUUCUGGCCCGGAGUGAAGCGCUCCGGGUCCCUUUUGGCCCAAUAGCAAUA------CGAACCGUAGCAAUGCGAACUGUUUGGCCCACUAGUUGGGCAC----CAAUCCC-CCCA ----.....(((((((((((...))))))))))).......(((((((((.....------(((((((((....))))....))))).....)).)))))))..----.......-.... ( -38.90) >DroYak_CAF1 19953 112 - 1 ----AUUUUGGCCCGGAGUGAUGCGCUCCGGGUCCCUUUUGGCCCAAUAGCAAUACGAAUACGAACCGUAGCAAUGCGAACUGUUUGGCCCACUAGCUGGGCAC----CAAUCCCCCCCA ----.....(((((((((((...))))))))))).......(((((.(((.....((((((.(...((((....))))..))))))).....)))..)))))..----............ ( -38.40) >DroAna_CAF1 22183 90 - 1 AACCGUCUCGGUCCGGAC---AUUGUUCCGGCAUCCUUCU---------------------CGAACUGUAGUA-UUCGAACUGUUUCGACUCCUUCAAAGGUCC----AUAUAUC-ACCA .(((.....((.(((((.---.....)))))...))...(---------------------((((...((((.-.....)))).)))))..........)))..----.......-.... ( -16.20) >consensus ____AUUUUGGCCCGGAGUGAUGCGCUCCGGGUCCCU_UUGGCUCAAUAGCAAUA______CGAACCGUAGCAAUCCGAACUGUUUGGCCCACCAGUUGGGCAC____CAAUCCC_CCCA .........((((((((((.....))))))))))...........................(((((.((..........)).)))))(((((.....))))).................. (-21.71 = -23.02 + 1.31)

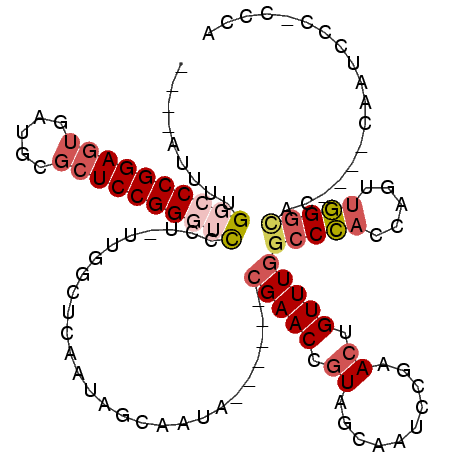
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:33 2006