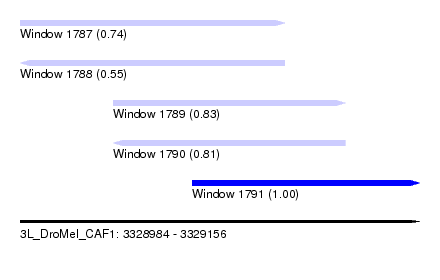
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,328,984 – 3,329,156 |
| Length | 172 |
| Max. P | 0.999807 |

| Location | 3,328,984 – 3,329,098 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.82 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -21.03 |
| Energy contribution | -21.51 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3328984 114 + 23771897 CAUAGAGUUAGUGGGAGAAGAGGGGACUUUGAGACCCUUUGGGAAG------AACCCUCGAUCUCUCUUAAUAAGCCAUCCCAAGCUAUUGCUAUUUUUAUGCGAAGACCUCUCUUCUCG (((((((.((((((((((.(((((..(((.....(((...))))))------..)))))..))))))......(((........)))...)))).))))))).(((((....)))))... ( -33.60) >DroSec_CAF1 16914 114 + 1 CAUAGAGUUAGUGGGAGAAGAGGGGACUUUGGGAACCUUUGGGAAU------AAACCUCGAUCUCUCUUAAUAAGCCAUCCCAAGCUAUUGCUAUUUUUAUGCGAAGACCUCUCUUCUCG ..............(((((((((((.((((((((..(((.((((..------...........)))).....)))...)))).(((....)))..........)))).))))))))))). ( -31.92) >DroSim_CAF1 9678 113 + 1 CAUAGAGUUUGUGGGAGAAGAGGGGACUUUGGGAACCU-UGGGAAG------AAACCUCGAUCUCUCUUAAUAAGCCAUCCCAAGCUAUUGCUAUUUUUAUGCGAAGACCUCUCUUCUCG (((((...))))).(((((((((((.((((((((..((-((..(((------(............))))..))))...)))).(((....)))..........)))).))))))))))). ( -35.40) >DroEre_CAF1 17308 105 + 1 UAUAGGCUACGUAAGAAAAGCGAGAGCUUUGGGAACCUUUU-AA--------AAACCUCGACUACUCUUAAUAAGCCAUCCCAAGCU------AUUUUUAUGCGAAGACCUCUCUUUUCG ...(((((.((((.(((((.....(((((.((((..(((((-((--------...............)))).)))...)))))))))------.))))).)))).)).)))......... ( -20.36) >DroYak_CAF1 12284 108 + 1 UGUAGGCUAAGAUGGAAAAGAGA-AGCUUUGGGA-CCUUUU-AAAGAAAAAAAAACUUCACUU---AAUAAUAAGCCAUCCCAAGCU------AUUUUUAUGCGAAGAGCUCUCUUUUCG ..............(((((((((-(((((.((((-......-...(((........))).(((---(....))))...)))))))))------........((.....))))))))))). ( -24.80) >consensus CAUAGAGUUAGUGGGAGAAGAGGGGACUUUGGGAACCUUUGGGAAG______AAACCUCGAUCUCUCUUAAUAAGCCAUCCCAAGCUAUUGCUAUUUUUAUGCGAAGACCUCUCUUCUCG ..............(((((((((((.((((((((..(((.................................)))...))))..((...............)))))).))))))))))). (-21.03 = -21.51 + 0.48)

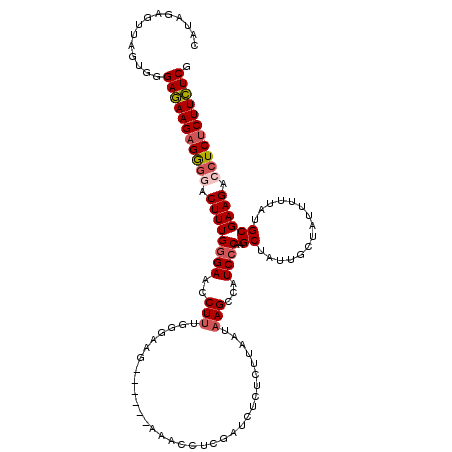
| Location | 3,328,984 – 3,329,098 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.82 |
| Mean single sequence MFE | -29.37 |
| Consensus MFE | -17.13 |
| Energy contribution | -16.65 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3328984 114 - 23771897 CGAGAAGAGAGGUCUUCGCAUAAAAAUAGCAAUAGCUUGGGAUGGCUUAUUAAGAGAGAUCGAGGGUU------CUUCCCAAAGGGUCUCAAAGUCCCCUCUUCUCCCACUAACUCUAUG .((((((((.((.(((.((........(((....))).......)).........((((((..(((..------...)))....)))))).))).)).)))))))).............. ( -32.66) >DroSec_CAF1 16914 114 - 1 CGAGAAGAGAGGUCUUCGCAUAAAAAUAGCAAUAGCUUGGGAUGGCUUAUUAAGAGAGAUCGAGGUUU------AUUCCCAAAGGUUCCCAAAGUCCCCUCUUCUCCCACUAACUCUAUG .((((((((.((.(((............((....))((((((.(((((.....((....)).))))).------..)))))).........))).)).)))))))).............. ( -29.00) >DroSim_CAF1 9678 113 - 1 CGAGAAGAGAGGUCUUCGCAUAAAAAUAGCAAUAGCUUGGGAUGGCUUAUUAAGAGAGAUCGAGGUUU------CUUCCCA-AGGUUCCCAAAGUCCCCUCUUCUCCCACAAACUCUAUG .((((((((.((.(((...........(((....)))(((((..((((.....(((((((....))))------)))....-)))))))))))).)).)))))))).............. ( -32.60) >DroEre_CAF1 17308 105 - 1 CGAAAAGAGAGGUCUUCGCAUAAAAAU------AGCUUGGGAUGGCUUAUUAAGAGUAGUCGAGGUUU--------UU-AAAAGGUUCCCAAAGCUCUCGCUUUUCUUACGUAGCCUAUA .((((((.(((((....))........------(((((((((..((((.(((((((.........)))--------))-)).)))))))).)))))))).)))))).............. ( -23.70) >DroYak_CAF1 12284 108 - 1 CGAAAAGAGAGCUCUUCGCAUAAAAAU------AGCUUGGGAUGGCUUAUUAUU---AAGUGAAGUUUUUUUUUCUUU-AAAAGG-UCCCAAAGCU-UCUCUUUUCCAUCUUAGCCUACA .(((((((((((.....))........------((((((((((.(((((....)---))))((((......))))...-.....)-)))).)))))-))))))))).............. ( -28.90) >consensus CGAGAAGAGAGGUCUUCGCAUAAAAAUAGCAAUAGCUUGGGAUGGCUUAUUAAGAGAGAUCGAGGUUU______CUUCCCAAAGGUUCCCAAAGUCCCCUCUUCUCCCACUAACUCUAUG .((((((((.((.(((((...............((((......)))).............)))))..................((...)).....)).)))))))).............. (-17.13 = -16.65 + -0.48)

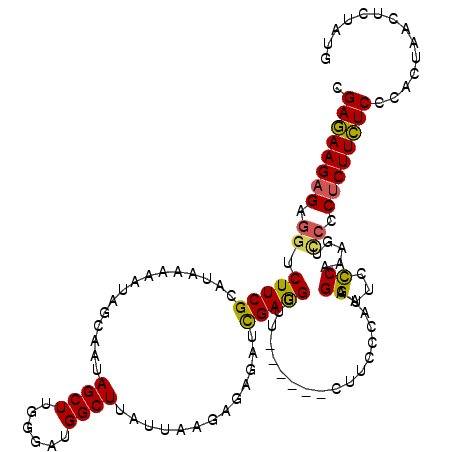
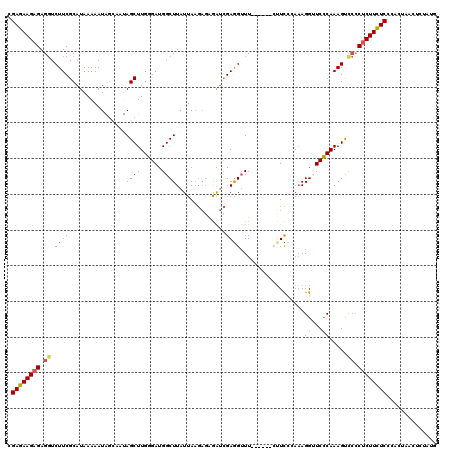
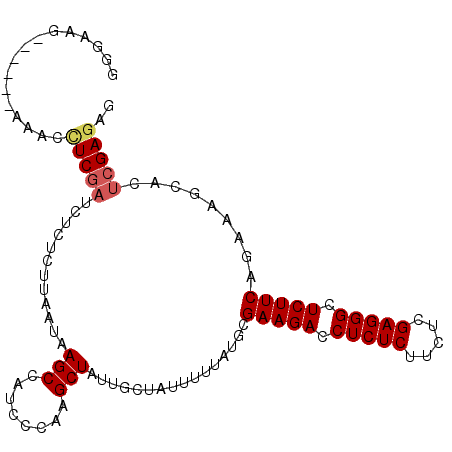
| Location | 3,329,024 – 3,329,124 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 83.87 |
| Mean single sequence MFE | -22.62 |
| Consensus MFE | -17.12 |
| Energy contribution | -17.60 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3329024 100 + 23771897 GGGAAG------AACCCUCGAUCUCUCUUAAUAAGCCAUCCCAAGCUAUUGCUAUUUUUAUGCGAAGACCUCUCUUCUCGAGGGCUCUUCAGAAAGCACUCGAGAG ..((((------(.(((((((.............((.......(((....)))........))(((((....)))))))))))).)))))................ ( -26.86) >DroSec_CAF1 16954 100 + 1 GGGAAU------AAACCUCGAUCUCUCUUAAUAAGCCAUCCCAAGCUAUUGCUAUUUUUAUGCGAAGACCUCUCUUCUCGAGGGCUCUUCAGAAAGCACUCGAGAG ......------....(((((.((.(((......((.......(((....)))........))(((((.(((((.....))))).)))))))).))...))))).. ( -23.16) >DroSim_CAF1 9717 100 + 1 GGGAAG------AAACCUCGAUCUCUCUUAAUAAGCCAUCCCAAGCUAUUGCUAUUUUUAUGCGAAGACCUCUCUUCUCGAGGGCUCUUCAGAAAGCACUCGAGAG ..((((------(..((((((.............((.......(((....)))........))(((((....)))))))))))..)))))................ ( -24.66) >DroEre_CAF1 17348 91 + 1 U-AA--------AAACCUCGACUACUCUUAAUAAGCCAUCCCAAGCU------AUUUUUAUGCGAAGACCUCUCUUUUCGAGGGCUCUUCAAAAACCACUCGACAG .-..--------.....((((............(((........)))------..........(((((.(((((.....))))).))))).........))))... ( -16.80) >DroYak_CAF1 12322 96 + 1 U-AAAGAAAAAAAAACUUCACUU---AAUAAUAAGCCAUCCCAAGCU------AUUUUUAUGCGAAGAGCUCUCUUUUCGAGGGCUCUUCAAAAACCACUCGAGAG .-...(((........))).(((---.......(((........)))------..........(((((((((((.....)))))))))))...........))).. ( -21.60) >consensus GGGAAG______AAACCUCGAUCUCUCUUAAUAAGCCAUCCCAAGCUAUUGCUAUUUUUAUGCGAAGACCUCUCUUCUCGAGGGCUCUUCAGAAAGCACUCGAGAG ................(((((............(((........)))................(((((.(((((.....))))).))))).........))))).. (-17.12 = -17.60 + 0.48)

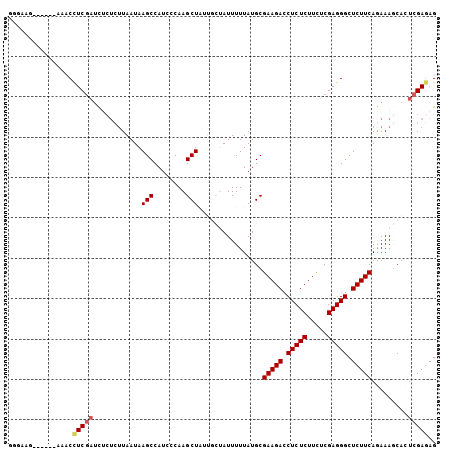
| Location | 3,329,024 – 3,329,124 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 83.87 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -19.64 |
| Energy contribution | -21.56 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3329024 100 - 23771897 CUCUCGAGUGCUUUCUGAAGAGCCCUCGAGAAGAGAGGUCUUCGCAUAAAAAUAGCAAUAGCUUGGGAUGGCUUAUUAAGAGAGAUCGAGGGUU------CUUCCC (((((((...(((((((((((.(((((.....))).)))))))((........(((....))).......))......)))))).)))))))..------...... ( -32.46) >DroSec_CAF1 16954 100 - 1 CUCUCGAGUGCUUUCUGAAGAGCCCUCGAGAAGAGAGGUCUUCGCAUAAAAAUAGCAAUAGCUUGGGAUGGCUUAUUAAGAGAGAUCGAGGUUU------AUUCCC .((((((...(((((((((((.(((((.....))).)))))))((........(((....))).......))......)))))).))))))...------...... ( -29.96) >DroSim_CAF1 9717 100 - 1 CUCUCGAGUGCUUUCUGAAGAGCCCUCGAGAAGAGAGGUCUUCGCAUAAAAAUAGCAAUAGCUUGGGAUGGCUUAUUAAGAGAGAUCGAGGUUU------CUUCCC .((((((...(((((((((((.(((((.....))).)))))))((........(((....))).......))......)))))).))))))...------...... ( -29.96) >DroEre_CAF1 17348 91 - 1 CUGUCGAGUGGUUUUUGAAGAGCCCUCGAAAAGAGAGGUCUUCGCAUAAAAAU------AGCUUGGGAUGGCUUAUUAAGAGUAGUCGAGGUUU--------UU-A ...((((((.(((((((((((.(((((.....))).)))))))....))))))------.))))))(((.((((.....)))).))).......--------..-. ( -23.20) >DroYak_CAF1 12322 96 - 1 CUCUCGAGUGGUUUUUGAAGAGCCCUCGAAAAGAGAGCUCUUCGCAUAAAAAU------AGCUUGGGAUGGCUUAUUAUU---AAGUGAAGUUUUUUUUUCUUU-A .((((((((.(((((((((((((.(((.....))).)))))))....))))))------.))))))))..(((((....)---))))((((......))))...-. ( -30.50) >consensus CUCUCGAGUGCUUUCUGAAGAGCCCUCGAGAAGAGAGGUCUUCGCAUAAAAAUAGCAAUAGCUUGGGAUGGCUUAUUAAGAGAGAUCGAGGUUU______CUUCCC .((((((...(((((((((((.(.(((.....))).).)))))................((((......)))).....)))))).))))))............... (-19.64 = -21.56 + 1.92)

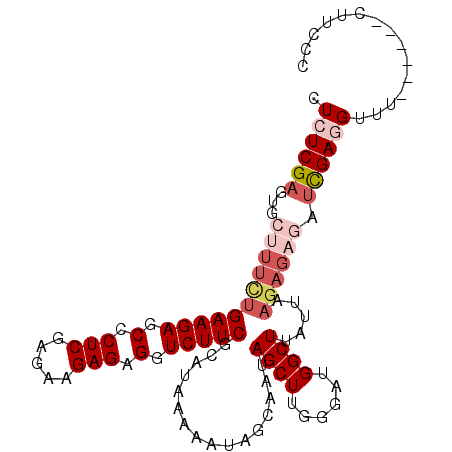
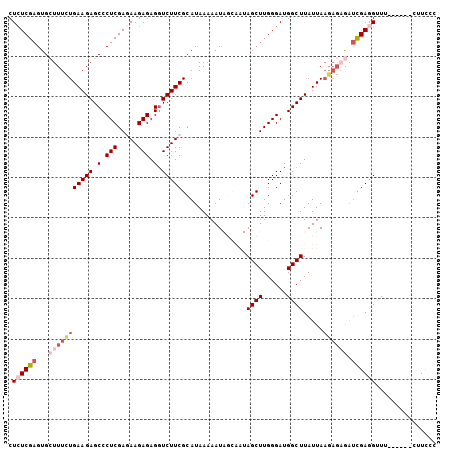
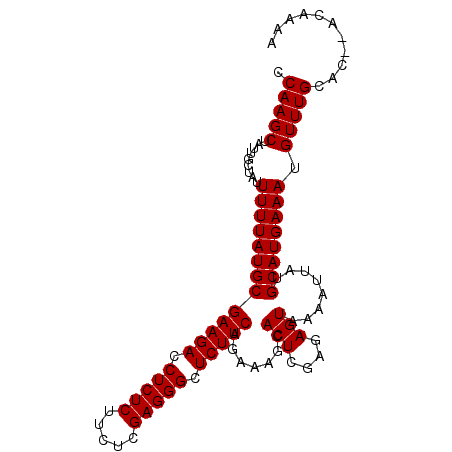
| Location | 3,329,058 – 3,329,156 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 92.46 |
| Mean single sequence MFE | -25.82 |
| Consensus MFE | -22.30 |
| Energy contribution | -22.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.86 |
| SVM decision value | 4.13 |
| SVM RNA-class probability | 0.999807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3329058 98 + 23771897 CCAAGCUAUUGCUAUUUUUAUGCGAAGACCUCUCUUCUCGAGGGCUCUUCAGAAAGCACUCGAGAGUAAAAUUAUGCAUGAAAUGUUUGCAC--ACAAAA ....((....((...(((((((((.(((....)))(((((((.(((........))).))))))).........))))))))).))..))..--...... ( -24.60) >DroSec_CAF1 16988 98 + 1 CCAAGCUAUUGCUAUUUUUAUGCGAAGACCUCUCUUCUCGAGGGCUCUUCAGAAAGCACUCGAGAGUAAAAUUAUGCAUGAAAUGUUUGCAC--ACAAAA ....((....((...(((((((((.(((....)))(((((((.(((........))).))))))).........))))))))).))..))..--...... ( -24.60) >DroSim_CAF1 9751 98 + 1 CCAAGCUAUUGCUAUUUUUAUGCGAAGACCUCUCUUCUCGAGGGCUCUUCAGAAAGCACUCGAGAGUAAAAUUAUGCAUGAAAUGUUUGCAC--ACAAAA ....((....((...(((((((((.(((....)))(((((((.(((........))).))))))).........))))))))).))..))..--...... ( -24.60) >DroEre_CAF1 17379 92 + 1 CCAAGCU------AUUUUUAUGCGAAGACCUCUCUUUUCGAGGGCUCUUCAAAAACCACUCGACAGUAAAAUUAUGCAUGAAAUGUUUGCAC--ACAAAA .(((((.------..(((((((((((((.(((((.....))))).))))).......(((....)))........)))))))).)))))...--...... ( -24.50) >DroYak_CAF1 12358 94 + 1 CCAAGCU------AUUUUUAUGCGAAGAGCUCUCUUUUCGAGGGCUCUUCAAAAACCACUCGAGAGUAAAAUUAUGCAUGAAACGUUUGCACACACAAAA .(((((.------..(((((((((((((((((((.....))))))))))).......(((....)))........)))))))).)))))........... ( -30.80) >consensus CCAAGCUAUUGCUAUUUUUAUGCGAAGACCUCUCUUCUCGAGGGCUCUUCAGAAAGCACUCGAGAGUAAAAUUAUGCAUGAAAUGUUUGCAC__ACAAAA .(((((.........(((((((((((((.(((((.....))))).))))).......(((....)))........)))))))).)))))........... (-22.30 = -22.30 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:25 2006