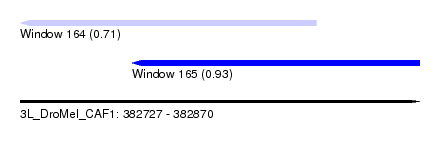
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 382,727 – 382,870 |
| Length | 143 |
| Max. P | 0.934045 |

| Location | 382,727 – 382,833 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.68 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -18.70 |
| Energy contribution | -19.78 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 382727 106 - 23771897 AAAAACUUUUUAGCUAAUGAAAGUCAAACUACUUUCGUU-------------GCUGGU-GGCAUCUUUCAAUUAGAAAACAUGCUUGGCUAAGUAGGAAAACUCUAAAGGGACAAGGACA .....((.((((((((((((((((......)))))))))-------------......-(((((.((((.....))))..))))).))))))).)).....(((....)))......... ( -21.80) >DroSec_CAF1 36821 119 - 1 AAAAACUUUUUAGCUAAUGAAAGUCAAACUACUUUCGUUGGCAGUCGCACAUGCUGGU-GGCAUCUUUCAAUUAGAAAACAUGCUUGGCUAAGUAGGAAAACUCUAAAGGGACAUGGGCA ............((((((((((((......))))))))))))....((.((((.((((-(((((.((((.....))))..)))))..))))..........(((....))).)))).)). ( -30.40) >DroSim_CAF1 37575 119 - 1 AAAAACUUUUUAGCUAAUGAAAGUCAAACUACUUUCGUUGGCAGUCGCACAUGCUGGU-GGCAUCUUUCAAUUAGAAAACCUGCUUGGCUAAGUAGGAAAACUCUAAAGGGACAUGGGCA ............((((((((((((......)))))))))))).........((((.((-(...((((.....((((...(((((((....))))))).....))))))))..))).)))) ( -33.50) >DroEre_CAF1 40248 101 - 1 AAAAACUUUUUAGCUAAUGGAAGUCAAACUACUUUCGUUGGC------------UGGU-GGCAUCUUUCAAUUAGAAAAUCCGCUUGGCUAAGUAGGAAAACUCUAAAGGGA------CA .........(((((((((((((((......))))))))))))------------))).-.......(((..(((((...(((((((....)))).)))....)))))..)))------.. ( -24.70) >DroYak_CAF1 38788 107 - 1 AAAAACUUUUUAGCUAAUGGAAGUCAAACUACU-GCAUUGGC------------AGUCCCACAUCUUUUAAUUAGAAAAUCUGCUUGGCUAAGUAGGAAAACUCUAAAGGGACAAGGACA .....((((...(((((((..(((......)))-.)))))))------------.(((((...(((.......)))...(((((((....)))))))...........)))))))))... ( -26.00) >consensus AAAAACUUUUUAGCUAAUGAAAGUCAAACUACUUUCGUUGGC__________GCUGGU_GGCAUCUUUCAAUUAGAAAACCUGCUUGGCUAAGUAGGAAAACUCUAAAGGGACAAGGACA .....(((((((((((((((((((......)))))))))))).....................................(((((((....))))))).......)))))))......... (-18.70 = -19.78 + 1.08)

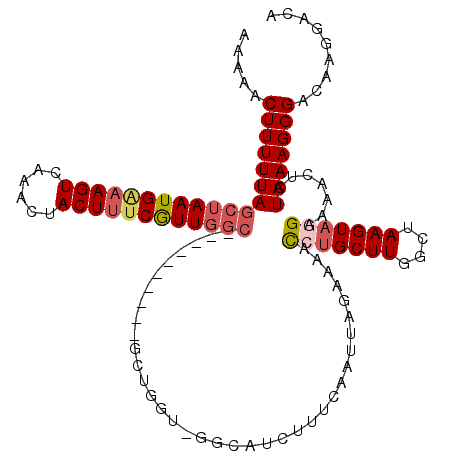
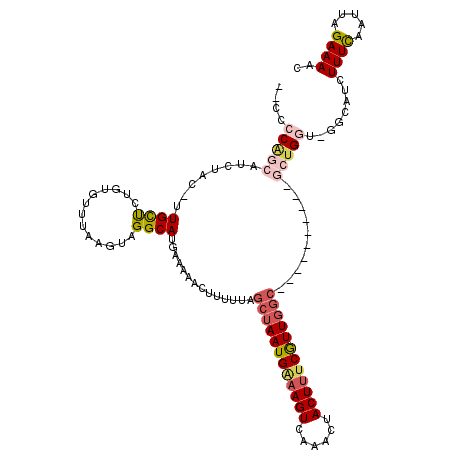
| Location | 382,767 – 382,870 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.70 |
| Mean single sequence MFE | -28.15 |
| Consensus MFE | -15.12 |
| Energy contribution | -16.64 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.934045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 382767 103 - 23771897 --CCCCAGCAUCUAC-UUGUUCUGCGUUUAAGUAGGCAUGAAAAACUUUUUAGCUAAUGAAAGUCAAACUACUUUCGUU-------------GCUGGU-GGCAUCUUUCAAUUAGAAAAC --(((((((..((((-(((.........)))))))((.(((((....))))))).(((((((((......)))))))))-------------))))).-))....((((.....)))).. ( -25.90) >DroSec_CAF1 36861 116 - 1 --CCCCAGCAUCUAC-UUGCUCUGUGUUUAAGUAGGCAUGAAAAACUUUUUAGCUAAUGAAAGUCAAACUACUUUCGUUGGCAGUCGCACAUGCUGGU-GGCAUCUUUCAAUUAGAAAAC --(((((((((((((-((((.....))..))))))((.(((((....)))))((((((((((((......))))))))))))....))..))))))).-))....((((.....)))).. ( -35.30) >DroSim_CAF1 37615 117 - 1 --CCACAGCAUCUAUUUUGCUCUGUGUUUAAGUAGGCAUGAAAAACUUUUUAGCUAAUGAAAGUCAAACUACUUUCGUUGGCAGUCGCACAUGCUGGU-GGCAUCUUUCAAUUAGAAAAC --(((((((((((((((.((.....))..))))))((.(((((....)))))((((((((((((......))))))))))))....))..))))).))-))....((((.....)))).. ( -31.80) >DroEre_CAF1 40282 106 - 1 CUCCCCGCCAUUUGC-UUGCCCAGUGUUUAUGUAGGCAUAAAAAACUUUUUAGCUAAUGGAAGUCAAACUACUUUCGUUGGC------------UGGU-GGCAUCUUUCAAUUAGAAAAU ......(((((.(((-((((...........)))))))............((((((((((((((......))))))))))))------------))))-)))...((((.....)))).. ( -30.70) >DroYak_CAF1 38828 106 - 1 GUCCCCAGCAUUUCC-UUGCUUAUUGUUUAAUUAGGCAUAAAAAACUUUUUAGCUAAUGGAAGUCAAACUACU-GCAUUGGC------------AGUCCCACAUCUUUUAAUUAGAAAAU ......((((.....-.)))).....(((((((((((.(((((....))))))))....((((.......(((-((....))------------))).......)))))))))))).... ( -17.04) >consensus __CCCCAGCAUCUAC_UUGCUCUGUGUUUAAGUAGGCAUGAAAAACUUUUUAGCUAAUGAAAGUCAAACUACUUUCGUUGGC__________GCUGGU_GGCAUCUUUCAAUUAGAAAAC ....(((((........((((.............))))..............((((((((((((......))))))))))))..........)))))........((((.....)))).. (-15.12 = -16.64 + 1.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:53 2006