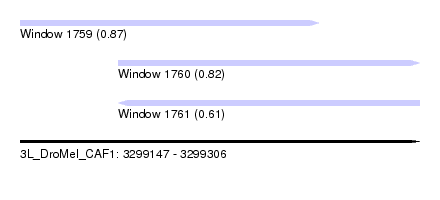
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,299,147 – 3,299,306 |
| Length | 159 |
| Max. P | 0.869098 |

| Location | 3,299,147 – 3,299,266 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.22 |
| Mean single sequence MFE | -41.52 |
| Consensus MFE | -27.81 |
| Energy contribution | -28.00 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3299147 119 + 23771897 AACCAGAAGAACAUUCUAAUA-AUGGCUCAAAUUAAAGGAUUGUUUGCUCUCCUCGCUGUGGUGACCAUUGUCCUAAUGGUGGCCAACUCGGCUUCGGCCGUGGAUUGCCCAUCUGGAAG ..(((((.(((((.(((..((-((.......))))..))).)))))((..(((.(....((((.(((((((...))))))).))))....(((....)))).)))..))...)))))... ( -41.60) >DroSim_CAF1 5093 119 + 1 AACCAGAAGAAUAUUCUAAAA-AUGGCUCAAAUUAAAGGAUUGUUUGCUCUCCUCGCUGUGGUGACCAUUGUCCUAAUGGUGGCCAACUCGGCUUCGGCCGUGGAUUGCCCAUCUGGAAG ..((((((((....)))....-..(((.........((((..(....)..))))(((..((((.(((((((...))))))).))))....(((....))))))....)))..)))))... ( -38.00) >DroEre_CAF1 5001 119 + 1 AACCAGAAGCUCAUUACAAAA-AUGGCUCAGAUUAAAGGCUUGUUCGCUGUGCUCGCUGUGGUGGCCAUUGUCCUAAUGGCCGCCAACUCGUCUUCGGCUGUGGAUUGCCAGUCUGGAAC ..((((((((.((((.....)-)))))).........(((..((((((...(((((...((((((((((((...))))))))))))...)).....))).)))))).)))..)))))... ( -44.70) >DroYak_CAF1 4305 120 + 1 UACCAAGACUUAAUAGUAAAACAUGGCUCAGAUGAAAGGCUUGUUCGCUGUGCUCGCUGUGGUGGCCAUUGUCCUUUUGGCCGCCGACUCGUCUUCGGCCGUGGAUUGCCUAUCUGGAAC ........((..(((((((..(((((((.((((((..(((..((.......))..))).(((((((((.........)))))))))..))))))..)))))))..))).))))..))... ( -41.80) >consensus AACCAGAAGAACAUUCUAAAA_AUGGCUCAAAUUAAAGGAUUGUUCGCUCUCCUCGCUGUGGUGACCAUUGUCCUAAUGGCCGCCAACUCGGCUUCGGCCGUGGAUUGCCCAUCUGGAAC ..(((((...............((((((..................((.......))..(((((((((((.....)))))))))))..........))))))((....))..)))))... (-27.81 = -28.00 + 0.19)

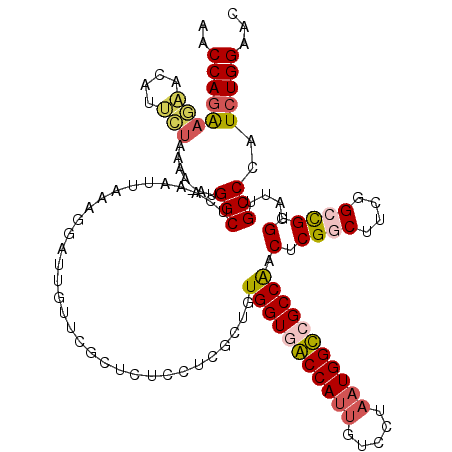
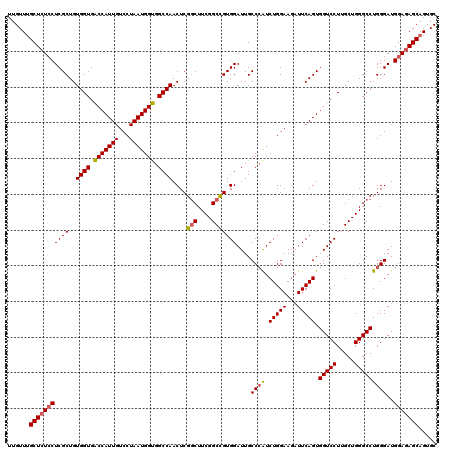
| Location | 3,299,186 – 3,299,306 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -53.37 |
| Consensus MFE | -42.90 |
| Energy contribution | -43.90 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3299186 120 + 23771897 UUGUUUGCUCUCCUCGCUGUGGUGACCAUUGUCCUAAUGGUGGCCAACUCGGCUUCGGCCGUGGAUUGCCCAUCUGGAAGAUUCAGUGGUCCUUGCUGGGCCUGGGAUGGAGAGCAGUGC ....(((((((((((((..((((.(((((((...))))))).))))....(((....)))))))....((((.(((((...))))).(((((.....)))))))))..)))))))))... ( -54.40) >DroSim_CAF1 5132 120 + 1 UUGUUUGCUCUCCUCGCUGUGGUGACCAUUGUCCUAAUGGUGGCCAACUCGGCUUCGGCCGUGGAUUGCCCAUCUGGAAGAUUCAGUGGUCCUUGCUGGGCCUGGGAUGGAGAGCAGUGC ....(((((((((((((..((((.(((((((...))))))).))))....(((....)))))))....((((.(((((...))))).(((((.....)))))))))..)))))))))... ( -54.40) >DroEre_CAF1 5040 120 + 1 UUGUUCGCUGUGCUCGCUGUGGUGGCCAUUGUCCUAAUGGCCGCCAACUCGUCUUCGGCUGUGGAUUGCCAGUCUGGAACUUUCAGUGGUCCUUGCUGGGCCUGGGAUGGAGAGCAGUGC .....((((((.(((....((((((((((((...))))))))))))...(((((..(((.........(((((..(((.((......)))))..))))))))..)))))))).)))))). ( -51.30) >consensus UUGUUUGCUCUCCUCGCUGUGGUGACCAUUGUCCUAAUGGUGGCCAACUCGGCUUCGGCCGUGGAUUGCCCAUCUGGAAGAUUCAGUGGUCCUUGCUGGGCCUGGGAUGGAGAGCAGUGC ......(((((((......((((.(((((((...))))))).)))).(((((.((((((...((((..(..(((.....)))...)..))))..)))))).)))))..)))))))..... (-42.90 = -43.90 + 1.00)

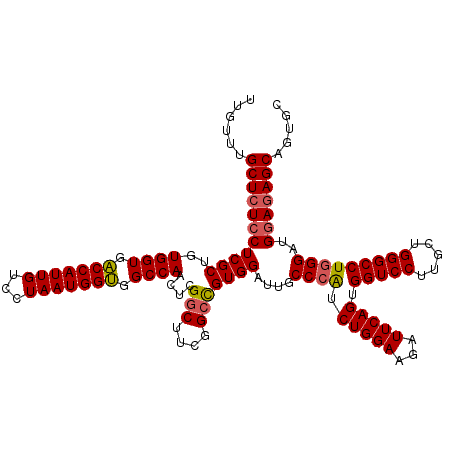
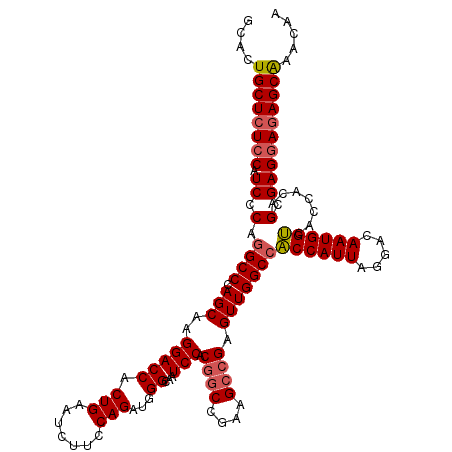
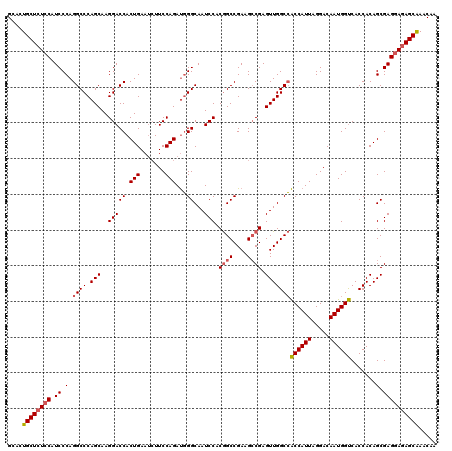
| Location | 3,299,186 – 3,299,306 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -40.80 |
| Consensus MFE | -32.76 |
| Energy contribution | -33.77 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3299186 120 - 23771897 GCACUGCUCUCCAUCCCAGGCCCAGCAAGGACCACUGAAUCUUCCAGAUGGGCAAUCCACGGCCGAAGCCGAGUUGGCCACCAUUAGGACAAUGGUCACCACAGCGAGGAGAGCAAACAA ....((((((((.((.(.((((.(((..(((((((((.......))).)))....))).((((....)))).)))))))((((((.....)))))).......).))))))))))..... ( -43.70) >DroSim_CAF1 5132 120 - 1 GCACUGCUCUCCAUCCCAGGCCCAGCAAGGACCACUGAAUCUUCCAGAUGGGCAAUCCACGGCCGAAGCCGAGUUGGCCACCAUUAGGACAAUGGUCACCACAGCGAGGAGAGCAAACAA ....((((((((.((.(.((((.(((..(((((((((.......))).)))....))).((((....)))).)))))))((((((.....)))))).......).))))))))))..... ( -43.70) >DroEre_CAF1 5040 120 - 1 GCACUGCUCUCCAUCCCAGGCCCAGCAAGGACCACUGAAAGUUCCAGACUGGCAAUCCACAGCCGAAGACGAGUUGGCGGCCAUUAGGACAAUGGCCACCACAGCGAGCACAGCGAACAA ((..(((((.........((.((.....)).)).(((............((((........)))).........(((.(((((((.....))))))).)))))).)))))..))...... ( -35.00) >consensus GCACUGCUCUCCAUCCCAGGCCCAGCAAGGACCACUGAAUCUUCCAGAUGGGCAAUCCACGGCCGAAGCCGAGUUGGCCACCAUUAGGACAAUGGUCACCACAGCGAGGAGAGCAAACAA ....((((((((.((.(.((((.(((..(((((.(((.......)))...))...))).((((....)))).)))))))((((((.....)))))).......).))))))))))..... (-32.76 = -33.77 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:56 2006