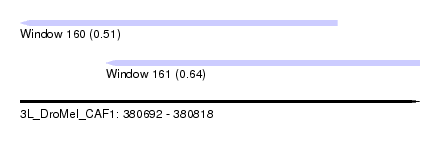
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 380,692 – 380,818 |
| Length | 126 |
| Max. P | 0.637449 |

| Location | 380,692 – 380,792 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 77.53 |
| Mean single sequence MFE | -18.39 |
| Consensus MFE | -11.29 |
| Energy contribution | -12.60 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
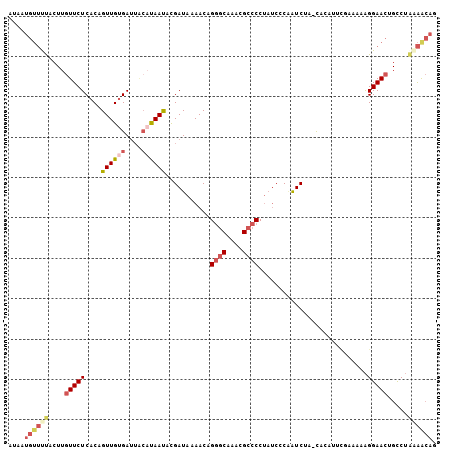
>3L_DroMel_CAF1 380692 100 - 23771897 -----GUUUUACUUGUUCUCACAGUUGUGAUUACAUAAUACGAUAAAACAGGGCAAACGCCCCUUUCCCAAUCUA-UACAUUCGAAAAAGGAACUGCCUAAAACAG -----((((((...(((((....((((((....))))))...........((((....)))).............-.............)))))....)))))).. ( -17.80) >DroSec_CAF1 34789 105 - 1 AUAAUGUUUUACUUGUUCUCACAGUUGUGAUUGCAUAACUCGAUAAAACAGGCCAAACGCCCCUAUCCCAAUCUA-UACAUUCGAAAAAGGAACUGCCUAAAACAG ....(((((((...(((((...(((((((....))))))).((((.....(((.....)))..))))........-.............)))))....))))))). ( -17.90) >DroSim_CAF1 35534 105 - 1 AUAAUGUUUUACUUGUUCUCACAGUUGUGAUUACAUAACUCGAUAAAACAGGGCAAACGCCCCUAUCCCAAUCUA-CACAUUCGAAAAAGGAACUGCCUAAAACAG ....(((((((...(((((...(((((((....))))))).((((.....((((....)))).))))........-.............)))))....))))))). ( -21.30) >DroEre_CAF1 36207 105 - 1 AUAAUGUUCUCCUUGUUCUCACAGUUGUGAUUACAUAAUACGAUAAAACCGGGCAAACGCCCCUAUCCCAAUCUA-CCCAUUUACAAAAGGAACUGUCUAGAGCAG ....((((((...(((..((((....))))..)))......((((.....((((....))))...(((.......-.............)))..)))).)))))). ( -20.55) >DroYak_CAF1 36658 100 - 1 UUAAUGUUUUCCUUGUUCUCACAGUUGUGA-----UAAUACGAUAAAACCGGGCAAACGCCCCUAUCCCUAUCUA-UAGAUUUACAAAAGGAACUGUCUUCAACAG ........(((((((((.((((....))))-----.)))..((((.....((((....)))).......))))..-...........))))))((((.....)))) ( -18.90) >DroAna_CAF1 31175 99 - 1 AGAAUC--CCGAUACUUCUCACAGUUGCGAUUACACAAUAUGAUAAAACAGAGCAAACGCCCCAUCAACGGUCAAACGCA-----UAAGGGAAAAGUCAGAGAAAC ......--.......(((((...((((.(....).)))).((((........((....))(((.....((......))..-----...)))....))))))))).. ( -13.90) >consensus AUAAUGUUUUACUUGUUCUCACAGUUGUGAUUACAUAAUACGAUAAAACAGGGCAAACGCCCCUAUCCCAAUCUA_CACAUUCGAAAAAGGAACUGCCUAAAACAG ....((((((....(((((....((((((....))))))...........((((....))))...........................))))).....)))))). (-11.29 = -12.60 + 1.31)

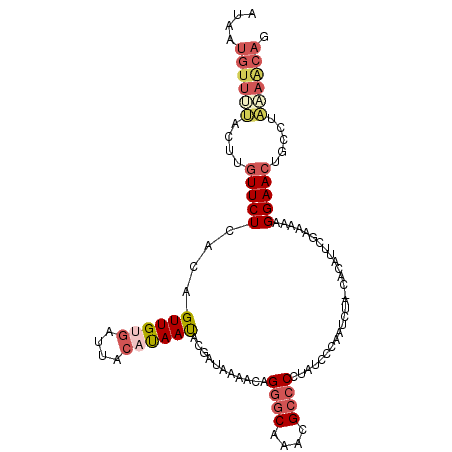
| Location | 380,719 – 380,818 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 71.06 |
| Mean single sequence MFE | -21.11 |
| Consensus MFE | -8.80 |
| Energy contribution | -9.36 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.637449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
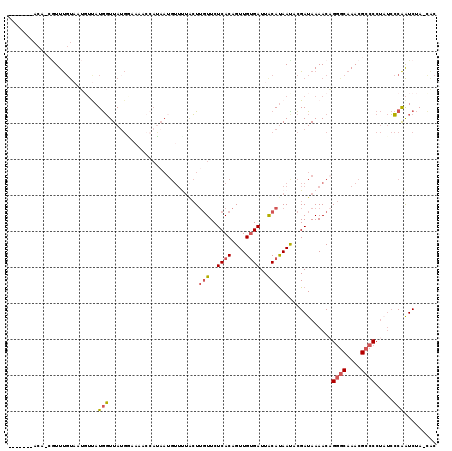
>3L_DroMel_CAF1 380719 99 - 23771897 -------ACAGCGUUUGUAAUGUUAUGGUUAUG------------GUUUUACUUGUUCUCACAGUUGUGAUUACAUAAUACGAUAAAACAGGGCAAACGCCCCUUUCCCAAUCUA-UAC -------...((((((((..((((....((((.------------((.(((..(((..((((....))))..)))))).)).))))))))..))))))))...............-... ( -24.50) >DroSec_CAF1 34816 109 - 1 -------AC--GGUUUGUAAUUUUAUGGUUAUGAAAAACCAUAAUGUUUUACUUGUUCUCACAGUUGUGAUUGCAUAACUCGAUAAAACAGGCCAAACGCCCCUAUCCCAAUCUA-UAC -------..--(((((((....((((((((......))))))))((((.....(((..((((....))))..)))......))))..))))))).....................-... ( -22.70) >DroSim_CAF1 35561 109 - 1 -------AC--GGUUUGUAAUGUUAUGGUUAUGGAAAACCAUAAUGUUUUACUUGUUCUCACAGUUGUGAUUACAUAACUCGAUAAAACAGGGCAAACGCCCCUAUCCCAAUCUA-CAC -------..--.((((((((((((((((((......))))))))))))...((((((.....(((((((....)))))))......)))))))))))).................-... ( -26.80) >DroEre_CAF1 36234 118 - 1 AUAAAAAACAACAUUUGUUAGGUCAUGGUUAAGGAAACCCAUAAUGUUCUCCUUGUUCUCACAGUUGUGAUUACAUAAUACGAUAAAACCGGGCAAACGCCCCUAUCCCAAUCUA-CCC ......((((.....)))).(((...((((((((((((.......))).)))))((..((((....))))..))............))))((((....))))............)-)). ( -22.50) >DroAna_CAF1 31197 113 - 1 CAAAAAAAUAACCUUCCC----ACAUAUUUAUCCUUACCCAGAAUC--CCGAUACUUCUCACAGUUGCGAUUACACAAUAUGAUAAAACAGAGCAAACGCCCCAUCAACGGUCAAACGC ..........(((.....----.....((((((.........((((--.((((..........)))).)))).........)))))).....((....)).........)))....... ( -9.07) >consensus _______ACA_CGUUUGUAAUGUUAUGGUUAUGGAAAACCAUAAUGUUUUACUUGUUCUCACAGUUGUGAUUACAUAAUACGAUAAAACAGGGCAAACGCCCCUAUCCCAAUCUA_CAC .........................(((.........................(((..((((....))))..)))...............((((....)))).....)))......... ( -8.80 = -9.36 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:48 2006