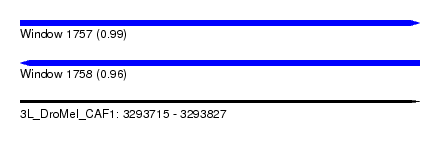
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,293,715 – 3,293,827 |
| Length | 112 |
| Max. P | 0.987575 |

| Location | 3,293,715 – 3,293,827 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.97 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -24.66 |
| Energy contribution | -25.46 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
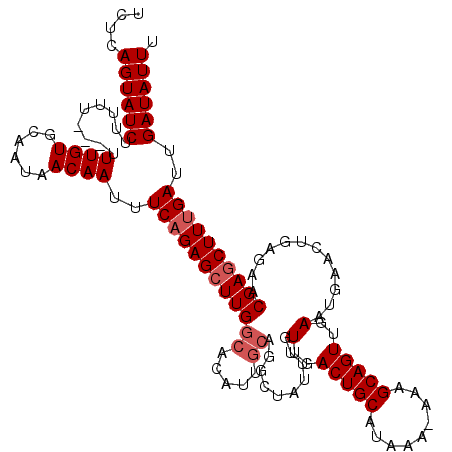
>3L_DroMel_CAF1 3293715 112 + 23771897 UCUCAGUAUCUUUUUU-UUUUGUGCAAUAACAAUUUCAGAGCUUGGC-------AGGCUAUUUGUUGACUGCAUAAAAAAAGCAGUUGAAUGAACUGAGAACGAGCUUUGAUUGAUAUUU ....((((((......-..((((......))))..((((((((((.(-------((..(((((...((((((.........)))))))))))..)))....))))))))))..)))))). ( -31.20) >DroSec_CAF1 23345 118 + 1 UCUCAGUAUCUUUUUUGUUUUGUGCAAUAACAAUUUCAGAGCUUGGCACAUUGCAGUCUAUUUGUUGACUGCAUAAA--AAGCAGUUGAAUGAACUGAGAACGAGCUUUGAUUGAUAUUU ....((((((.........((((......))))..((((((((((......(((((((........)))))))....--...(((((.....)))))....))))))))))..)))))). ( -33.70) >DroSim_CAF1 24257 116 + 1 UCUCAGUAUCUUUUU---UUUGUGCAAUAACAAUUUCAGAGCUUGGCACAUUGCAGUCUAUUUGUUGACUGCAUAAA-AAAGCAGUUGAAUGAACUGAGAACGAGCUUUGAUUGAUAUUU ....((((((.....---.((((......))))..((((((((((......(((((((........)))))))....-....(((((.....)))))....))))))))))..)))))). ( -33.70) >DroEre_CAF1 25494 115 + 1 UCUCAGUAUCUUUU----UUUGUGCAAUAACAAUUUCAGAGCUUGGCACAUUGCAAGCUAUUUGUUGACUGCAUAAA-AAAGCAGUUGAAUGAACUGAGAACGAGCUUUGAUUGAUAUUU (((((((...((((----((((((((.((((((......((((((........))))))..))))))..))))))))-))))((......)).))))))).................... ( -29.70) >DroYak_CAF1 26113 115 + 1 UCUCAGUAUCUUUU----UUUGUGCAAUAACAAUUUCCGAGCUUGGCACAUUGCAGGCUAUUUGUUGACUGCAUAAA-GAGGCAGUUGAAUGAACUGAGAACGAACUUUGAUUGAUAUUU (((((((...((((----((((((((.((((((......((((((........))))))..))))))..))))))))-))))((......)).))))))).................... ( -29.10) >consensus UCUCAGUAUCUUUUU___UUUGUGCAAUAACAAUUUCAGAGCUUGGCACAUUGCAGGCUAUUUGUUGACUGCAUAAA_AAAGCAGUUGAAUGAACUGAGAACGAGCUUUGAUUGAUAUUU ....((((((.........((((......))))..((((((((((((.....))..........(..(((((.........)))))..)............))))))))))..)))))). (-24.66 = -25.46 + 0.80)

| Location | 3,293,715 – 3,293,827 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.97 |
| Mean single sequence MFE | -24.35 |
| Consensus MFE | -18.86 |
| Energy contribution | -19.46 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
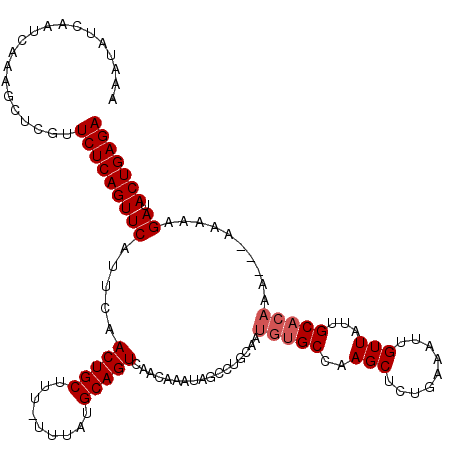
>3L_DroMel_CAF1 3293715 112 - 23771897 AAAUAUCAAUCAAAGCUCGUUCUCAGUUCAUUCAACUGCUUUUUUUAUGCAGUCAACAAAUAGCCU-------GCCAAGCUCUGAAAUUGUUAUUGCACAAAA-AAAAAAGAUACUGAGA ....................(((((((........((..(((((((.((((((.(((((..(((..-------.....)))......))))))))))).))))-)))..))..))))))) ( -19.80) >DroSec_CAF1 23345 118 - 1 AAAUAUCAAUCAAAGCUCGUUCUCAGUUCAUUCAACUGCUU--UUUAUGCAGUCAACAAAUAGACUGCAAUGUGCCAAGCUCUGAAAUUGUUAUUGCACAAAACAAAAAAGAUACUGAGA ....................(((((((...........(((--(((.(((((((........))))))).(((((..(((.........)))...))))).....))))))..))))))) ( -26.82) >DroSim_CAF1 24257 116 - 1 AAAUAUCAAUCAAAGCUCGUUCUCAGUUCAUUCAACUGCUUU-UUUAUGCAGUCAACAAAUAGACUGCAAUGUGCCAAGCUCUGAAAUUGUUAUUGCACAAA---AAAAAGAUACUGAGA ....................(((((((...........((((-(((.(((((((........))))))).(((((..(((.........)))...))))).)---))))))..))))))) ( -27.72) >DroEre_CAF1 25494 115 - 1 AAAUAUCAAUCAAAGCUCGUUCUCAGUUCAUUCAACUGCUUU-UUUAUGCAGUCAACAAAUAGCUUGCAAUGUGCCAAGCUCUGAAAUUGUUAUUGCACAAA----AAAAGAUACUGAGA ....................(((((((...........((((-(((.((((((.(((((..(((((((.....).))))))......)))))))))))..))----)))))..))))))) ( -25.22) >DroYak_CAF1 26113 115 - 1 AAAUAUCAAUCAAAGUUCGUUCUCAGUUCAUUCAACUGCCUC-UUUAUGCAGUCAACAAAUAGCCUGCAAUGUGCCAAGCUCGGAAAUUGUUAUUGCACAAA----AAAAGAUACUGAGA ....................(((((((((.....(((((...-.....))))).........((..(((((...((......))..)))))....)).....----....)).))))))) ( -22.20) >consensus AAAUAUCAAUCAAAGCUCGUUCUCAGUUCAUUCAACUGCUUU_UUUAUGCAGUCAACAAAUAGCCUGCAAUGUGCCAAGCUCUGAAAUUGUUAUUGCACAAA___AAAAAGAUACUGAGA ....................(((((((((.....(((((.........))))).................(((((..(((.........)))...)))))..........)).))))))) (-18.86 = -19.46 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:53 2006