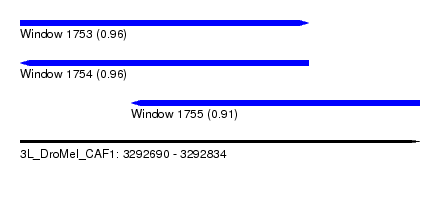
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,292,690 – 3,292,834 |
| Length | 144 |
| Max. P | 0.960680 |

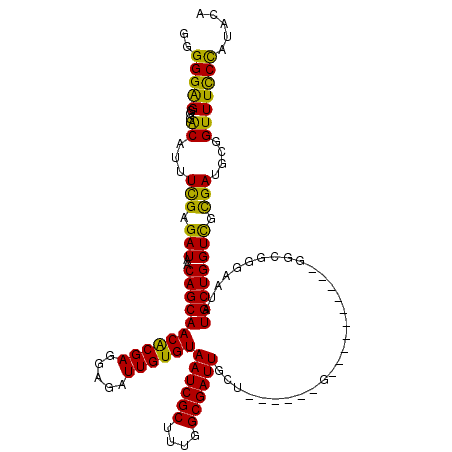
| Location | 3,292,690 – 3,292,794 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.73 |
| Mean single sequence MFE | -38.98 |
| Consensus MFE | -29.08 |
| Energy contribution | -27.72 |
| Covariance contribution | -1.36 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3292690 104 + 23771897 GGGGGGGGGUACGUUUCGAGAUAACAGCAACACGAGGAGAUUGUGUAAUCGCUUUGGCGAUUGCU------G----------GGCGGGAAUAUGCUGGUCGCGAUGCGGUUUCCCAUACA (...(((((..(((.(((.(((..((((.((((((.....))))))((((((....)))))))))------)----------((((......)))).))).))).)))..)))))...). ( -39.10) >DroSec_CAF1 22405 104 + 1 GGGGGAGGGGACAUUUCGAGAUAACAGCAACGCGAGGAGAUUGUGUAAUCGCUUUGGCGAUUGCU------G----------GGCGGGCAUAUGCUGGUCGCGAUGCGGUUUCCCAUACA (((..((....)..............(((.(((...........((((((((....)))))))).------.----------(((.(((....))).)))))).)))..)..)))..... ( -38.40) >DroSim_CAF1 23219 104 + 1 GGGGGAGGGGACAUUUCGAGAUAACAGCAACACGAGGAGAUUGUGUAAUCGCUUUGGCGAUUGCU------G----------GGCGGGAAUAUGCUGGUCGCGAUGCGGUUUCCCAUACA (((..((....)...(((.(((..((((.((((((.....))))))((((((....)))))))))------)----------((((......)))).))).))).....)..)))..... ( -35.90) >DroEre_CAF1 24431 116 + 1 --GGGAG-GGGCAGUUUGAGAUAACAGCAACACGAGGAGAUUGUGUAAUCGCGUUGGCGAUUGUUGGGGUUGGGAUUGUUGGGGCGGC-AUAUGCUGGUUGCGAUGCUGUUUCCCAUAUA --(((((-.((((((((..(((..((((.((((((.....))))))((((((....))))))))))..))).))))))))...(((((-((.(((.....)))))))))))))))..... ( -43.40) >DroYak_CAF1 25103 111 + 1 --GGGAG-GGGCAGUUCGAGAUAACAGCAACACGAGGAGAUUGUGUAAUCGCUUUGGCGAUUGUU------GGGAUUGCUGGGGCGGGAAAAUGCUGGUCGUGAUGCGGUUUUCUAUACA --(..((-..(((...(((.....(((((((((((.....))))))..((((((..((((((...------..))))))..)))))).....))))).)))...)))..))..)...... ( -38.10) >consensus GGGGGAGGGGACAUUUCGAGAUAACAGCAACACGAGGAGAUUGUGUAAUCGCUUUGGCGAUUGCU______G__________GGCGGGAAUAUGCUGGUCGCGAUGCGGUUUCCCAUACA ..(((((...((...(((.(((..(((((((((((.....))))))((((((....))))))..............................)))))))).)))....)))))))..... (-29.08 = -27.72 + -1.36)

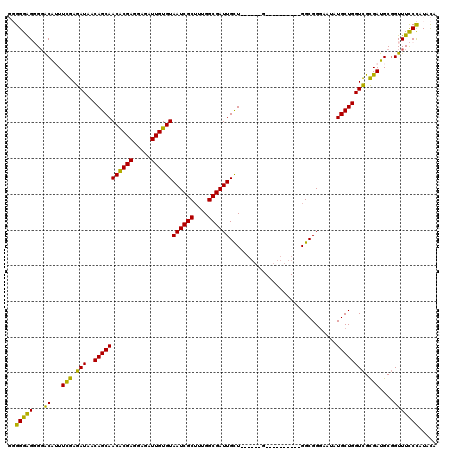
| Location | 3,292,690 – 3,292,794 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.73 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -20.92 |
| Energy contribution | -21.04 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3292690 104 - 23771897 UGUAUGGGAAACCGCAUCGCGACCAGCAUAUUCCCGCC----------C------AGCAAUCGCCAAAGCGAUUACACAAUCUCCUCGUGUUGCUGUUAUCUCGAAACGUACCCCCCCCC .((((((....))...(((.((.(((((..........----------.------...((((((....))))))((((.........)))))))))...)).)))...))))........ ( -23.70) >DroSec_CAF1 22405 104 - 1 UGUAUGGGAAACCGCAUCGCGACCAGCAUAUGCCCGCC----------C------AGCAAUCGCCAAAGCGAUUACACAAUCUCCUCGCGUUGCUGUUAUCUCGAAAUGUCCCCUCCCCC .....((((....((((..(((.(((((.((((..((.----------.------.))((((((....)))))).............))))))))).....)))..))))....)))).. ( -26.90) >DroSim_CAF1 23219 104 - 1 UGUAUGGGAAACCGCAUCGCGACCAGCAUAUUCCCGCC----------C------AGCAAUCGCCAAAGCGAUUACACAAUCUCCUCGUGUUGCUGUUAUCUCGAAAUGUCCCCUCCCCC .....((((....((((..(((...((........)).----------(------(((((((((....))))))((((.........)))).)))).....)))..))))....)))).. ( -25.20) >DroEre_CAF1 24431 116 - 1 UAUAUGGGAAACAGCAUCGCAACCAGCAUAU-GCCGCCCCAACAAUCCCAACCCCAACAAUCGCCAACGCGAUUACACAAUCUCCUCGUGUUGCUGUUAUCUCAAACUGCCC-CUCCC-- ....((((((((((((.(((.....((....-))........................((((((....)))))).............))).))))))).)))))........-.....-- ( -25.50) >DroYak_CAF1 25103 111 - 1 UGUAUAGAAAACCGCAUCACGACCAGCAUUUUCCCGCCCCAGCAAUCCC------AACAAUCGCCAAAGCGAUUACACAAUCUCCUCGUGUUGCUGUUAUCUCGAACUGCCC-CUCCC-- .....(((.(((.(((.(((((..((.(((.....((....))......------...((((((....))))))....)))))..))))).))).))).)))..........-.....-- ( -21.30) >consensus UGUAUGGGAAACCGCAUCGCGACCAGCAUAUUCCCGCC__________C______AGCAAUCGCCAAAGCGAUUACACAAUCUCCUCGUGUUGCUGUUAUCUCGAAAUGUCCCCUCCCCC ....((((((((.(((.(((((...((........)).....................((((((....))))))...........))))).))).))).)))))................ (-20.92 = -21.04 + 0.12)

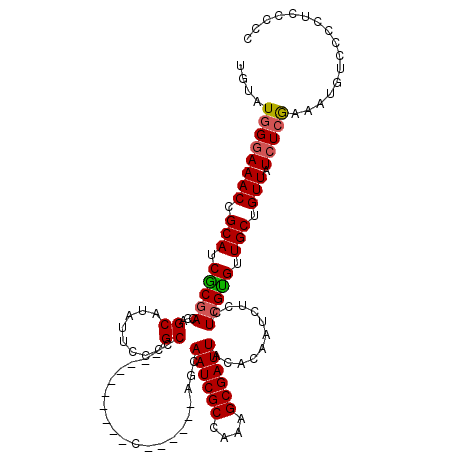
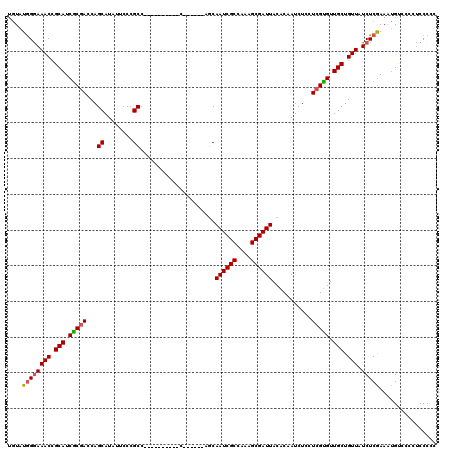
| Location | 3,292,730 – 3,292,834 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.10 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -18.16 |
| Energy contribution | -18.56 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3292730 104 - 23771897 ACCUAUGAUGCACUACACUCUGCGAUUGGACAGAACUCAUUGUAUGGGAAACCGCAUCGCGACCAGCAUAUUCCCGCC----------C------AGCAAUCGCCAAAGCGAUUACACAA ......(((((.........((((((.((......)).)))))).((....)))))))((.....((........)).----------.------.))((((((....))))))...... ( -25.10) >DroSec_CAF1 22445 104 - 1 ACCUAUGAUGCACUACACGCUACGAUUGGACAGAACCCAUUGUAUGGGAAACCGCAUCGCGACCAGCAUAUGCCCGCC----------C------AGCAAUCGCCAAAGCGAUUACACAA ......(((((.........((((((.((......)).)))))).((....)))))))(((..((.....))..))).----------.------.(.((((((....)))))).).... ( -28.30) >DroSim_CAF1 23259 104 - 1 ACCUAUGAUGCACUACACUCUACGAUUGGACAGAACCCAUUGUAUGGGAAACCGCAUCGCGACCAGCAUAUUCCCGCC----------C------AGCAAUCGCCAAAGCGAUUACACAA ......(((((.........((((((.((......)).)))))).((....)))))))((.....((........)).----------.------.))((((((....))))))...... ( -26.90) >DroEre_CAF1 24468 119 - 1 AAAUAUGAUGCACUAAACUCUGCGAUUGGAUUGAAUCGCCUAUAUGGGAAACAGCAUCGCAACCAGCAUAU-GCCGCCCCAACAAUCCCAACCCCAACAAUCGCCAACGCGAUUACACAA ......(((((..........((((((......)))))).......(....).)))))((.....))....-..........................((((((....))))))...... ( -23.10) >DroYak_CAF1 25140 114 - 1 AGCUAUGAUGCACUAAACUGUGCGAUUGGAUAGACUCCACUGUAUAGAAAACCGCAUCACGACCAGCAUUUUCCCGCCCCAGCAAUCCC------AACAAUCGCCAAAGCGAUUACACAA .(((.((((((......(((((((..((((.....)))).)))))))......))))))......((........))...)))......------...((((((....))))))...... ( -26.60) >consensus ACCUAUGAUGCACUACACUCUGCGAUUGGACAGAACCCAUUGUAUGGGAAACCGCAUCGCGACCAGCAUAUUCCCGCC__________C______AGCAAUCGCCAAAGCGAUUACACAA .....((((((.........((((((.((......)).)))))).((....))))))))......((........)).....................((((((....))))))...... (-18.16 = -18.56 + 0.40)

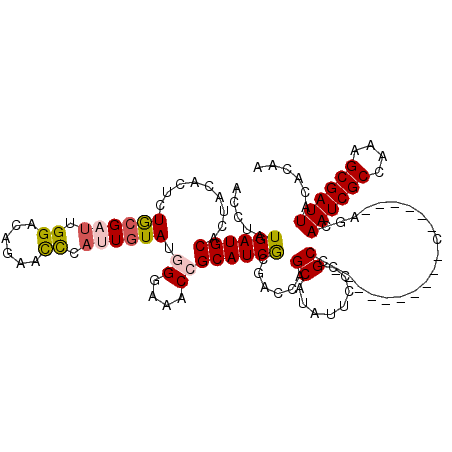
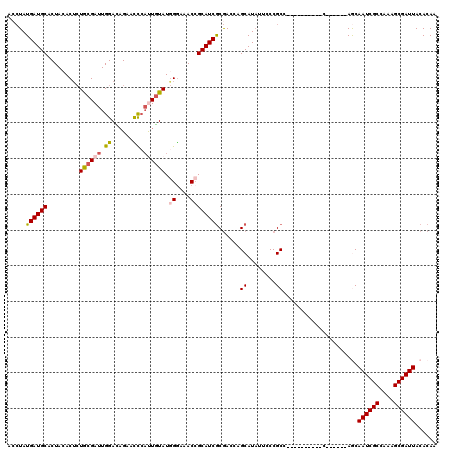
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:50 2006