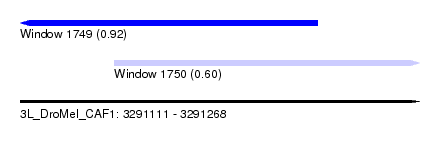
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,291,111 – 3,291,268 |
| Length | 157 |
| Max. P | 0.921352 |

| Location | 3,291,111 – 3,291,228 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.04 |
| Mean single sequence MFE | -30.99 |
| Consensus MFE | -24.84 |
| Energy contribution | -25.68 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
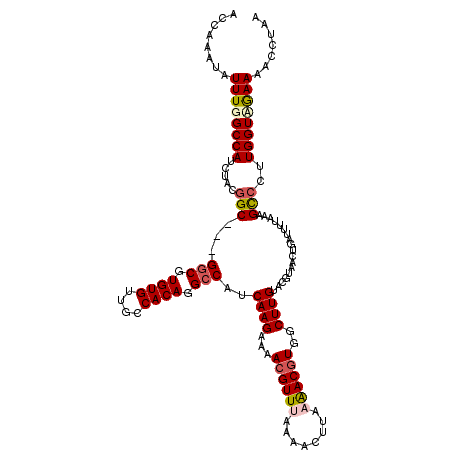
>3L_DroMel_CAF1 3291111 117 - 23771897 ACCAAAUAUUUGGCCAUCUACGGCGGCGGCGUGUGUUGCCACAGGCCAUCAAGAAAACGUUUAAAACUUAAAACGUGGCUUGUACGU---UUGUUUUAAAGCACUUGGUUGAAAACCUAA ........((..((((......((((((((....))))))((((((...((((...((((((........))))))..))))...))---))))......))...))))..))....... ( -33.10) >DroSec_CAF1 20862 117 - 1 ACCAAAUAUUUGGCCAUCUUCGGC---GGCGUGUGUUGCCACAGGCCAUCAAGAAAACGUUUAAAACUUAAAACGUGGCUUGUACGCAACUGAUUUUAAAGCCCUUGGUAGAAAACCUAA ............(((......)))---(((.((((....)))).)))((((((...((((((........))))))(((((.....(....)......)))))))))))........... ( -30.40) >DroSim_CAF1 21667 117 - 1 ACCAAAUAUUUGGCCAUCUACGGC---GGCGUGUGUUGCCACAGGCCAUCAAGAAAACGUUUAAAACUUAAAACGUGGCUUGUACGUAACUGAUUUUAAAGCCCUUGGUAGAAAACCUAA .((((....))))...(((((...---(((.((((....)))).)))..((((...((((((........))))))(((((.................))))))))))))))........ ( -31.23) >DroEre_CAF1 22969 109 - 1 ACCAAAUAUUUGGCCAUAUACGGC---GGCUUGUGUUGUCACAGGCCAUCAAGAAAACGUUUAA-------GACAUGGCUUGUACGUAACUGAAUUUG-AGUCCUUGGUCGAAAACAGAA ........((((((((.....(((---((((((((....)))))))).(((((...((((.(((-------(......)))).)))).......))))-))))..))))))))....... ( -30.60) >DroYak_CAF1 23589 110 - 1 ACCAAAUAUUUGGCCAUCUACGGC---GACUUGUGUUGUCACAGGCCAUCAAGUAAACGUUUAU-------AACGUGGCUUGUACGUAACUGAUUUUAAAGCCCUUGGUCAAAAACUAAA ........((((((((..((((((---(((....))))))(((((((((...((((....))))-------...)))))))))..))).((........))....))))))))....... ( -29.60) >consensus ACCAAAUAUUUGGCCAUCUACGGC___GGCGUGUGUUGCCACAGGCCAUCAAGAAAACGUUUAAAACUUAAAACGUGGCUUGUACGUAACUGAUUUUAAAGCCCUUGGUAGAAAACCUAA ........((((((((.....(((...(((.((((....)))).)))..((((...((((((........))))))..))))..................)))..))))))))....... (-24.84 = -25.68 + 0.84)

| Location | 3,291,148 – 3,291,268 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.81 |
| Mean single sequence MFE | -31.08 |
| Consensus MFE | -22.44 |
| Energy contribution | -22.72 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
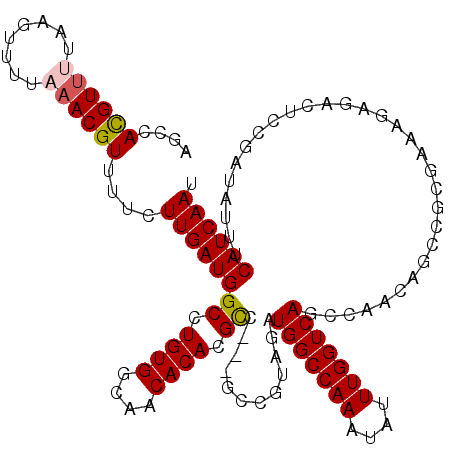
>3L_DroMel_CAF1 3291148 120 + 23771897 AGCCACGUUUUAAGUUUUAAACGUUUUCUUGAUGGCCUGUGGCAACACACGCCGCCGCCGUAGAUGGCCAAAUAUUUGGUCAGCCAACAGCCGCGAAAGAGACUCCGAUAUUUCAUCAAU ....((((((........))))))((((((...(((.((((....)))).)))((.((.((.(.(((((((....)))))))..).)).)).))..))))))....(((.....)))... ( -35.10) >DroSec_CAF1 20902 117 + 1 AGCCACGUUUUAAGUUUUAAACGUUUUCUUGAUGGCCUGUGGCAACACACGCC---GCCGAAGAUGGCCAAAUAUUUGGUCAGCCAACAGCCGCGAAAGAGACUCCGAUAUUUCAUCAAU .(((((((((........))))))..(((((.((((.((((....)))).)))---).).)))).))).(((((((.((((.((.....))..(....).))).).)))))))....... ( -31.60) >DroSim_CAF1 21707 117 + 1 AGCCACGUUUUAAGUUUUAAACGUUUUCUUGAUGGCCUGUGGCAACACACGCC---GCCGUAGAUGGCCAAAUAUUUGGUCAGCCAACAGCCGCGAAAGAGACUCCGAUAUUUCAUCAAU ....((((((........))))))....(((((((..((((....))))(((.---((.((.(.(((((((....)))))))..).)).)).))).................))))))). ( -32.20) >DroEre_CAF1 23008 99 + 1 AGCCAUGUC-------UUAAACGUUUUCUUGAUGGCCUGUGACAACACAAGCC---GCCGUAUAUGGCCAAAUAUUUGGUCAGCCA-----------AGAGACUUGGCUAUUUCAUCAAU (((((.(((-------((............(.((((.((((....)))).)))---).).....(((((((....)))))))....-----------.))))).)))))........... ( -31.10) >DroYak_CAF1 23629 110 + 1 AGCCACGUU-------AUAAACGUUUACUUGAUGGCCUGUGACAACACAAGUC---GCCGUAGAUGGCCAAAUAUUUGGUCAGCCAACAGCCGCGAAAGAGACUUUGAUAUUUCAUCAAU ....((((.-------....))))....(((((((...(((....)))(((((---((.((.(.(((((((....)))))))..).)).))..(....).))))).......))))))). ( -25.40) >consensus AGCCACGUUUUAAGUUUUAAACGUUUUCUUGAUGGCCUGUGGCAACACACGCC___GCCGUAGAUGGCCAAAUAUUUGGUCAGCCAACAGCCGCGAAAGAGACUCCGAUAUUUCAUCAAU ....((((((........))))))....((((((((.((((....)))).))............(((((((....)))))))...............................)))))). (-22.44 = -22.72 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:46 2006