| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,290,595 – 3,290,746 |
| Length | 151 |
| Max. P | 0.899633 |

| Location | 3,290,595 – 3,290,707 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 88.86 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -24.11 |
| Energy contribution | -26.30 |
| Covariance contribution | 2.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
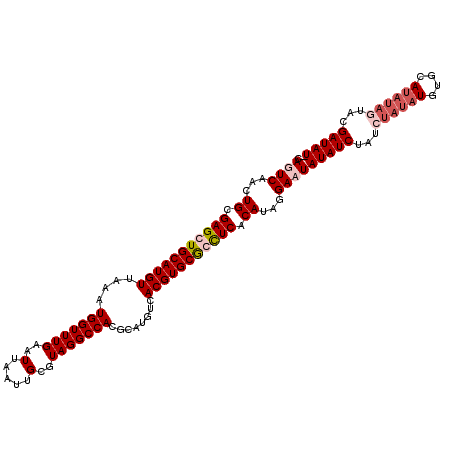
>3L_DroMel_CAF1 3290595 112 + 23771897 CAGUCAACUGCGAGCUGCAUGUUAAAUGGUUUGAAUUAAUUGCGUAGGCCACGCAUGUCACGUGCGCCUCACAUAGGACUAUAUCUAACUAUAUGUGCAUAUAGUAUGAUAU-- .((((...((.(((.(((((((....((((....))))..(((((.....)))))....))))))).))).))...))))(((((..(((((((....)))))))..)))))-- ( -30.10) >DroSec_CAF1 20327 114 + 1 CAGUCAACUGCGAGCUGCAUGUUAAAUGGUUUGAAUUAAUUGCGUAGGCCACGCAUGUCACGUGCGGCUCACAUAGGAAUAUAUCUAUCUAUAUGUGCAUAUAGUACGAUAUAU ...((...((.(((((((((((....((((....))))..(((((.....)))))....))))))))))).))...))(((((((...((((((....))))))...))))))) ( -33.60) >DroSim_CAF1 21143 102 + 1 CAGUCAACUGCGAGCUGCAUGUUAAAUGGUUUGAAUUAAUUGCUUAGGCCACGCAUGUCACGUGCGGCUCACAUAGGAAUAUAUCUA------------UAUAGUACGAUAUAU ...((...((.(((((((((((....((((((((.........))))))))........))))))))))).))...))(((((((..------------........))))))) ( -28.00) >DroEre_CAF1 22406 111 + 1 CAGUCAACUGUGAGCUGCAUGUUAAAUGGUUUGAAUUAAUUGCGUAGGCCACGCAUGUCACGUGCACUUCACAUAGGACUAUAUC-AUAUAUAUGUACAUAUAGUACGAUAU-- .((((...((((((.(((((((....((((....))))..(((((.....)))))....))))))).))))))...)))).....-..((((.(((((.....)))))))))-- ( -28.20) >consensus CAGUCAACUGCGAGCUGCAUGUUAAAUGGUUUGAAUUAAUUGCGUAGGCCACGCAUGUCACGUGCGCCUCACAUAGGAAUAUAUCUAUCUAUAUGUGCAUAUAGUACGAUAU__ .((((...((.(((((((((((....(((((((..(.....)..)))))))........))))))))))).))...))))(((((...((((((....))))))...))))).. (-24.11 = -26.30 + 2.19)

| Location | 3,290,595 – 3,290,707 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 88.86 |
| Mean single sequence MFE | -28.53 |
| Consensus MFE | -19.90 |
| Energy contribution | -22.77 |
| Covariance contribution | 2.88 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3290595 112 - 23771897 --AUAUCAUACUAUAUGCACAUAUAGUUAGAUAUAGUCCUAUGUGAGGCGCACGUGACAUGCGUGGCCUACGCAAUUAAUUCAAACCAUUUAACAUGCAGCUCGCAGUUGACUG --(((((..(((((((....)))))))..)))))((((...((((((((.(((((.....))))))))).)))).................(((.(((.....)))))))))). ( -32.90) >DroSec_CAF1 20327 114 - 1 AUAUAUCGUACUAUAUGCACAUAUAGAUAGAUAUAUUCCUAUGUGAGCCGCACGUGACAUGCGUGGCCUACGCAAUUAAUUCAAACCAUUUAACAUGCAGCUCGCAGUUGACUG (((((((...((((((....))))))...)))))))((...(((((((.(((.((....((((((...))))))..(((..........))))).))).)))))))...))... ( -29.30) >DroSim_CAF1 21143 102 - 1 AUAUAUCGUACUAUA------------UAGAUAUAUUCCUAUGUGAGCCGCACGUGACAUGCGUGGCCUAAGCAAUUAAUUCAAACCAUUUAACAUGCAGCUCGCAGUUGACUG (((((((........------------..)))))))((...(((((((.(((.((...(((..(((..(((....)))..)))...)))...)).))).)))))))...))... ( -24.20) >DroEre_CAF1 22406 111 - 1 --AUAUCGUACUAUAUGUACAUAUAUAU-GAUAUAGUCCUAUGUGAAGUGCACGUGACAUGCGUGGCCUACGCAAUUAAUUCAAACCAUUUAACAUGCAGCUCACAGUUGACUG --((((((((.(((((....))))))))-)))))((((...(((((..((((.((....((((((...))))))..(((..........))))).))))..)))))...)))). ( -27.70) >consensus __AUAUCGUACUAUAUGCACAUAUAGAUAGAUAUAGUCCUAUGUGAGCCGCACGUGACAUGCGUGGCCUACGCAAUUAAUUCAAACCAUUUAACAUGCAGCUCGCAGUUGACUG ..(((((...((((((....))))))...)))))((((...(((((((.(((.((....((((((...))))))..(((..........))))).))).)))))))...)))). (-19.90 = -22.77 + 2.88)

| Location | 3,290,635 – 3,290,746 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 85.40 |
| Mean single sequence MFE | -25.23 |
| Consensus MFE | -15.31 |
| Energy contribution | -18.12 |
| Covariance contribution | 2.81 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
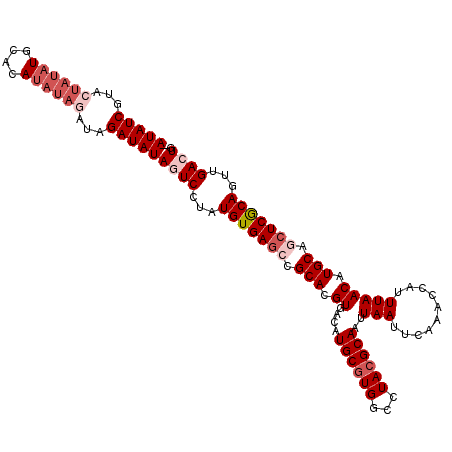
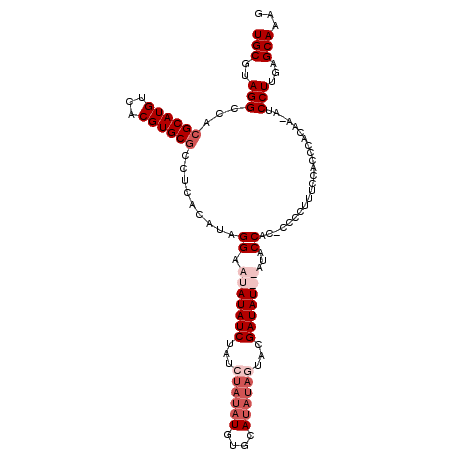
>3L_DroMel_CAF1 3290635 111 + 23771897 UGCGUAGGCCACGCAUGUCACGUGCGCCUCACAUAGGACUAUAUCUAACUAUAUGUGCAUAUAGUAUGAUAU--AUUCCACCCCCCUUUCCACCCACAA-GUCCUUGAGCAAAG (((((((((((((.......)))).)))).))...(((.((((((..(((((((....)))))))..)))))--).)))....................-........)))... ( -30.60) >DroSec_CAF1 20367 112 + 1 UGCGUAGGCCACGCAUGUCACGUGCGGCUCACAUAGGAAUAUAUCUAUCUAUAUGUGCAUAUAGUACGAUAUAUAUACCAC-CCCCUUUCCACCCACAA-AUCCUUGAGCAAAG (((((..(((.(((.......))).)))..))...((.(((((((...((((((....))))))...)))))))...))..-.................-........)))... ( -25.90) >DroSim_CAF1 21183 101 + 1 UGCUUAGGCCACGCAUGUCACGUGCGGCUCACAUAGGAAUAUAUCUA------------UAUAGUACGAUAUAUAUACCACACCCCUUUCCAGCCACAA-AUCCUUGAGCAAAG ((((((((....(((((...)))))((((.....(((........((------------((((......)))))).........)))....))))....-...))))))))... ( -23.73) >DroEre_CAF1 22446 110 + 1 UGCGUAGGCCACGCAUGUCACGUGCACUUCACAUAGGACUAUAUC-AUAUAUAUGUACAUAUAGUACGAUAU--ACGCCAA-CCCCUUUCCACCCACAACGUCCUUGAGCAAAG (((..(((.(..(((((...)))))..........((..((((((-...(((((....)))))....)))))--)..))..-..................).)))...)))... ( -20.70) >consensus UGCGUAGGCCACGCAUGUCACGUGCGCCUCACAUAGGAAUAUAUCUAUCUAUAUGUGCAUAUAGUACGAUAU__AUACCAC_CCCCUUUCCACCCACAA_AUCCUUGAGCAAAG (((..(((...((((((...)))))).........((.(((((((...((((((....))))))...)))))))...)).......................)))...)))... (-15.31 = -18.12 + 2.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:44 2006