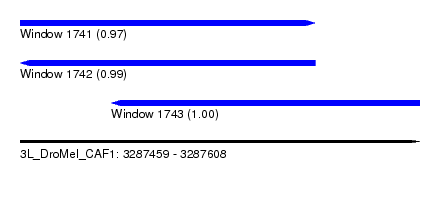
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,287,459 – 3,287,608 |
| Length | 149 |
| Max. P | 0.999969 |

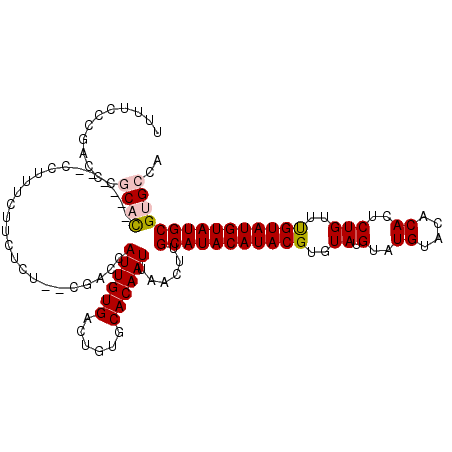
| Location | 3,287,459 – 3,287,569 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 92.69 |
| Mean single sequence MFE | -26.36 |
| Consensus MFE | -20.02 |
| Energy contribution | -20.50 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3287459 110 + 23771897 UUUUCCCGACCCGCAC------CCUUUCUUCUCUUUCGACCAUUGUGACUGUGCACAAUUAACUUGCAUACAUACGUGUAUGUAUGUACACACUCUGUUUGUAUGUAUGCGUGCCA ............((((------...................((((((......))))))......((((((((((((((((....))))))((...))..)))))))))))))).. ( -27.80) >DroSec_CAF1 17256 110 + 1 UUUUCCCGACCCUCCU------CCUUUCUUCUUUCACGACCAUUGUGACUGUGCACAAUUAACUUGCAUACAUACGUGUAUGUAUGUACACACUCUGUUUGUAUGUAUGCGUGCCA ................------............(((....((((((......))))))......((((((((((((((((....))))))((...))..)))))))))))))... ( -24.70) >DroSim_CAF1 18101 110 + 1 UUUUCCCGACCCUCCC------CCUUUCUUCUUUCACGACCAUUGUGACUGUGCACAAUUAACUUGCAUACAUACGCGUAUGUAUGUACACACUCUGUUUGUAUGUAUGCGUGCCA ................------...........((((((...)))))).((((((.........))))))..((((((((((((((.(((.....))).))))))))))))))... ( -22.80) >DroEre_CAF1 19280 114 + 1 UUUUCCCGACCCGCACCCGCCCCCUUUCUUCUCU--CGUCCAUUGUGACUGUGCACAAUUAACUUGCAUACAUACGUAUAUGUAUGUACACACUCUGUUUGUAUGUAUGCGUGCCA ............((((..................--.((..((((((......))))))..))..(((((((((((((((....)))))((.....))..)))))))))))))).. ( -27.50) >DroYak_CAF1 19875 108 + 1 UUUUCCCGACCCGCAC------CCUUUCUUCUCU--CGUCCAUUGUGACUGUGCACAAUUAACUUGCAUACAUACGUAUAUGUAUGUACACACUCUGUUCGUAUGUAUGCGUGCCA ............((((------............--.((..((((((......))))))..))..(((((((((((.(((.(..((....))..)))).))))))))))))))).. ( -29.00) >consensus UUUUCCCGACCCGCAC______CCUUUCUUCUCU__CGACCAUUGUGACUGUGCACAAUUAACUUGCAUACAUACGUGUAUGUAUGUACACACUCUGUUUGUAUGUAUGCGUGCCA ............((((.........................((((((......))))))......(((((((((((..((.(..((....))..)))..))))))))))))))).. (-20.02 = -20.50 + 0.48)

| Location | 3,287,459 – 3,287,569 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 92.69 |
| Mean single sequence MFE | -29.26 |
| Consensus MFE | -24.96 |
| Energy contribution | -24.72 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3287459 110 - 23771897 UGGCACGCAUACAUACAAACAGAGUGUGUACAUACAUACACGUAUGUAUGCAAGUUAAUUGUGCACAGUCACAAUGGUCGAAAGAGAAGAAAGG------GUGCGGGUCGGGAAAA .(((.((((..(.........((.((((((((((((((....))))))))(((.....))))))))).)).......((....)).......).------.)))).)))....... ( -31.00) >DroSec_CAF1 17256 110 - 1 UGGCACGCAUACAUACAAACAGAGUGUGUACAUACAUACACGUAUGUAUGCAAGUUAAUUGUGCACAGUCACAAUGGUCGUGAAAGAAGAAAGG------AGGAGGGUCGGGAAAA ...(((((((((((((.....(.((((((....)))))).)))))))))))..(...((((((......))))))...))))............------................ ( -25.70) >DroSim_CAF1 18101 110 - 1 UGGCACGCAUACAUACAAACAGAGUGUGUACAUACAUACGCGUAUGUAUGCAAGUUAAUUGUGCACAGUCACAAUGGUCGUGAAAGAAGAAAGG------GGGAGGGUCGGGAAAA ...(((((((((((((.......((((((....))))))..))))))))))..(...((((((......))))))...))))............------................ ( -26.30) >DroEre_CAF1 19280 114 - 1 UGGCACGCAUACAUACAAACAGAGUGUGUACAUACAUAUACGUAUGUAUGCAAGUUAAUUGUGCACAGUCACAAUGGACG--AGAGAAGAAAGGGGGCGGGUGCGGGUCGGGAAAA .(((.((((..(............(((((((((((......))))))))))).(((.((((((......)))))).))).--................)..)))).)))....... ( -30.80) >DroYak_CAF1 19875 108 - 1 UGGCACGCAUACAUACGAACAGAGUGUGUACAUACAUAUACGUAUGUAUGCAAGUUAAUUGUGCACAGUCACAAUGGACG--AGAGAAGAAAGG------GUGCGGGUCGGGAAAA ..(((((((((((((((......((((((....)))))).)))))))))))..(((.((((((......)))))).))).--............------))))............ ( -32.50) >consensus UGGCACGCAUACAUACAAACAGAGUGUGUACAUACAUACACGUAUGUAUGCAAGUUAAUUGUGCACAGUCACAAUGGUCG__AGAGAAGAAAGG______GUGCGGGUCGGGAAAA .(((..((((((((((.....(.((((((....)))))).)))))))))))..(...((((((......))))))...)...........................)))....... (-24.96 = -24.72 + -0.24)

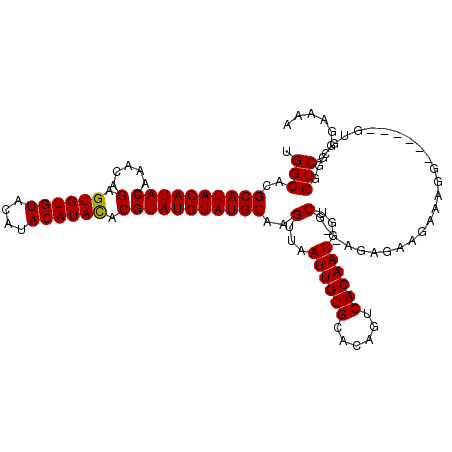
| Location | 3,287,493 – 3,287,608 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 96.20 |
| Mean single sequence MFE | -33.52 |
| Consensus MFE | -33.02 |
| Energy contribution | -32.78 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.99 |
| SVM decision value | 5.03 |
| SVM RNA-class probability | 0.999969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3287493 115 - 23771897 AAAUUCCUUUGCUUCUCC-UUUUUCCGCCUCAAUUGGGGGUGGCACGCAUACAUACAAACAGAGUGUGUACAUACAUACACGUAUGUAUGCAAGUUAAUUGUGCACAGUCACAAUG ..........((((....-.....(((((((.....)))))))...((((((((((.....(.((((((....)))))).)))))))))))))))..((((((......)))))). ( -33.20) >DroSec_CAF1 17290 115 - 1 AAAUUCCUUCGCUUCUCC-UUUUUCCGCCUCAAUUGGGGGUGGCACGCAUACAUACAAACAGAGUGUGUACAUACAUACACGUAUGUAUGCAAGUUAAUUGUGCACAGUCACAAUG ..........((((....-.....(((((((.....)))))))...((((((((((.....(.((((((....)))))).)))))))))))))))..((((((......)))))). ( -33.30) >DroSim_CAF1 18135 115 - 1 AAAUUCCUUCGCUUCUCC-UUUUUCCGCCUCAAUUGGGGGUGGCACGCAUACAUACAAACAGAGUGUGUACAUACAUACGCGUAUGUAUGCAAGUUAAUUGUGCACAGUCACAAUG ..........((((....-.....(((((((.....)))))))...((((((((((.......((((((....))))))..))))))))))))))..((((((......)))))). ( -33.90) >DroEre_CAF1 19318 116 - 1 AAAUUCCUUCGCUUUUCCCUGUUUCCGCCUCACUUGGGGGUGGCACGCAUACAUACAAACAGAGUGUGUACAUACAUAUACGUAUGUAUGCAAGUUAAUUGUGCACAGUCACAAUG ..........((((......((..(((((((.....))))))).))((((((((((.....(.((((((....)))))).)))))))))))))))..((((((......)))))). ( -32.20) >DroYak_CAF1 19907 116 - 1 AAAUUCCUUCGCUUUUCCCUGCUUCCGCCUCAAUUGGGGGUGGCACGCAUACAUACGAACAGAGUGUGUACAUACAUAUACGUAUGUAUGCAAGUUAAUUGUGCACAGUCACAAUG ..........((((.....((((.((.((......)).)).)))).(((((((((((......((((((....)))))).)))))))))))))))..((((((......)))))). ( -35.00) >consensus AAAUUCCUUCGCUUCUCC_UUUUUCCGCCUCAAUUGGGGGUGGCACGCAUACAUACAAACAGAGUGUGUACAUACAUACACGUAUGUAUGCAAGUUAAUUGUGCACAGUCACAAUG ..........((((..........(((((((.....)))))))...((((((((((.....(.((((((....)))))).)))))))))))))))..((((((......)))))). (-33.02 = -32.78 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:39 2006