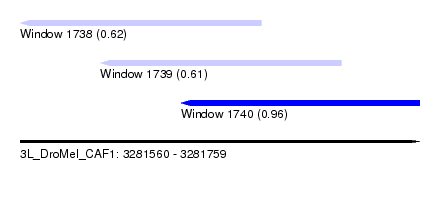
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,281,560 – 3,281,759 |
| Length | 199 |
| Max. P | 0.957472 |

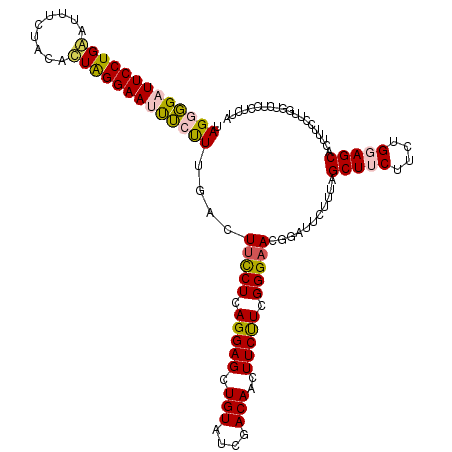
| Location | 3,281,560 – 3,281,680 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.58 |
| Mean single sequence MFE | -32.39 |
| Consensus MFE | -23.11 |
| Energy contribution | -23.54 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3281560 120 - 23771897 UAGGGGAUUCCUGAAUCUCUACAUUAGGAAUUUGUUUGACUUCCUCAGGAGCUGUAUCGACAACUUCUUCGGGAAUGGAUUCUUUAGCUUCUUCUGCAGCACUUUCCUUGGCCUCCUCUA (((((((..((..............(((((..((((....(((((.(((((.(((....))).)))))..)))))...........((.......))))))..))))).))..))))))) ( -28.56) >DroSec_CAF1 11534 120 - 1 UAGGGGAUUCCUGGACUUCUACACUAGGAAUCUCUUUGACUUCCUCAGGAGCUGUAUCGACAACUUCUUCGGGAACGGAUUCCUUAGCUUCUUCUGGAGCACUUUCCUCGGUCUCCUCUA (((((((..((.(((..........((((((((.......(((((.(((((.(((....))).)))))..))))).))))))))..(((((....)))))....)))..))..))))))) ( -38.80) >DroSim_CAF1 12385 120 - 1 UAGGGGUUUCCUGAAUUUCUACACUGGGAAUCUCUUUGACUUCCUCAGGAGCUGUAUCGACAACUUCCUCGGGAACGGAUUCCUUAGCUUCUUCUGGAGCACUUUCCUUGGUCUCCUCUA ((((((...((.(((..........((((((((.......(((((.(((((.(((....))).).)))).))))).))))))))..(((((....)))))...)))...))...)))))) ( -34.80) >DroEre_CAF1 11733 105 - 1 CAGGUGAUUCC---------------GGAACUGCCUUAACUUUCUCAGGAGAUGUAUCGACAACUUCUUGGGGUACGGUUUCUUCAGCUACUUGUGGAGCACUUUCCUUGGUCUCCUCAU .((.(((....---------------((((((((((((........(((((.(((....))).)))))))))))..)))))).))).))...((.((((.(((......))))))).)). ( -26.70) >DroYak_CAF1 12170 120 - 1 CAGGGGAUUCCUGGAUUUCUGCACUAGGAAUUUGUUUGACUUCCUCAGGAGAUGUAUCGACAAGUUCUUCGGGAACGGUUUCUUUAGCUUCUUCUGGAGCACUUUCCUCGACCUUCUCAA ..((((((((((((.........)))))))...........)))))..((((....((((.((((.((((((((..((((.....))))..)))))))).))))...))))...)))).. ( -33.10) >consensus UAGGGGAUUCCUGAAUUUCUACACUAGGAAUUUCUUUGACUUCCUCAGGAGCUGUAUCGACAACUUCUUCGGGAACGGAUUCUUUAGCUUCUUCUGGAGCACUUUCCUUGGUCUCCUCUA .(((((((((((((.........)))))))))))))....(((((.(((((.(((....)))..))))).)))))...........(((((....))))).................... (-23.11 = -23.54 + 0.43)

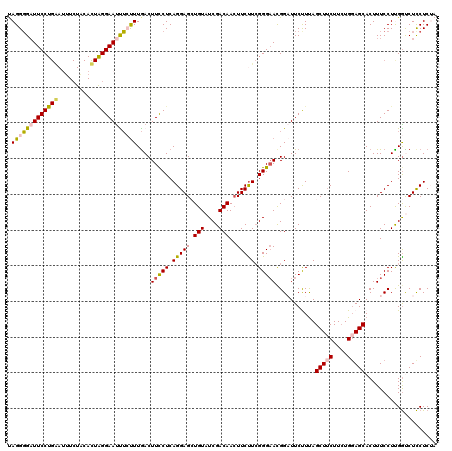
| Location | 3,281,600 – 3,281,720 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.58 |
| Mean single sequence MFE | -33.37 |
| Consensus MFE | -25.59 |
| Energy contribution | -25.62 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3281600 120 - 23771897 UCCUUGAUUUCUUCAGGAGAAGAUUCUUUAGGCUCCUCCAUAGGGGAUUCCUGAAUCUCUACAUUAGGAAUUUGUUUGACUUCCUCAGGAGCUGUAUCGACAACUUCUUCGGGAAUGGAU (((((((.....)))(((((((..((....(((((((.....(((((((....))))))).....(((((.((....)).))))).))))))).....))...)))))))))))...... ( -33.10) >DroSec_CAF1 11574 120 - 1 UCCUUGACUUCUUCAGGAGAAGAUUCUUUAGUCUCCUCCAUAGGGGAUUCCUGGACUUCUACACUAGGAAUCUCUUUGACUUCCUCAGGAGCUGUAUCGACAACUUCUUCGGGAACGGAU (((.........(((((((.((((......)))).))))..(((((((((((((.........)))))))))))))))).(((((.(((((.(((....))).)))))..))))).))). ( -39.70) >DroSim_CAF1 12425 120 - 1 UCCUUGACUUCUUCAGGAGAAGAUUCUUUAGGCUCCUCCAUAGGGGUUUCCUGAAUUUCUACACUGGGAAUCUCUUUGACUUCCUCAGGAGCUGUAUCGACAACUUCCUCGGGAACGGAU .(((.((..(((((....))))).))...))).(((..((.((((((((((..............)))))))))).))..(((((.(((((.(((....))).).)))).))))).))). ( -34.14) >DroEre_CAF1 11773 105 - 1 UCCUUGAUCUCUUCAGGAGAAGUUUCUUUGGGCUCCUCUACAGGUGAUUCC---------------GGAACUGCCUUAACUUUCUCAGGAGAUGUAUCGACAACUUCUUGGGGUACGGUU .((...((((((....((((((((.....((((...(((...((.....))---------------)))...)))).))).))))).))))))(((((..(((....))).))))))).. ( -25.60) >DroYak_CAF1 12210 120 - 1 UCCUUAAUUUCUUCAGGAGCAGUUUCUUUGGGCUCCUCUACAGGGGAUUCCUGGAUUUCUGCACUAGGAAUUUGUUUGACUUCCUCAGGAGAUGUAUCGACAAGUUCUUCGGGAACGGUU ((((.((....)).))))((((..(((..((..(((((....)))))..)).)))...))))...(((((((((((..((((((...))))..))...)))))))))))((....))... ( -34.30) >consensus UCCUUGAUUUCUUCAGGAGAAGAUUCUUUAGGCUCCUCCAUAGGGGAUUCCUGAAUUUCUACACUAGGAAUUUCUUUGACUUCCUCAGGAGCUGUAUCGACAACUUCUUCGGGAACGGAU .((............((((.((((......)))).))))..(((((((((((((.........)))))))))))))....(((((.(((((.(((....)))..))))).))))).)).. (-25.59 = -25.62 + 0.03)

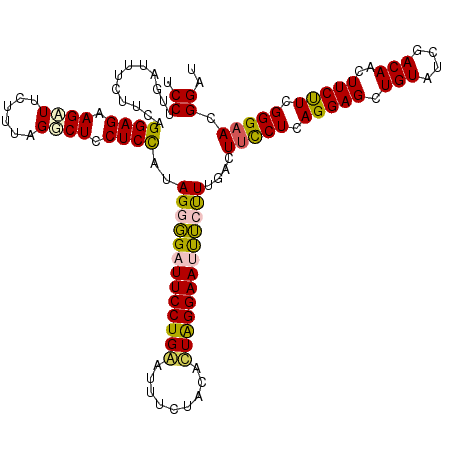
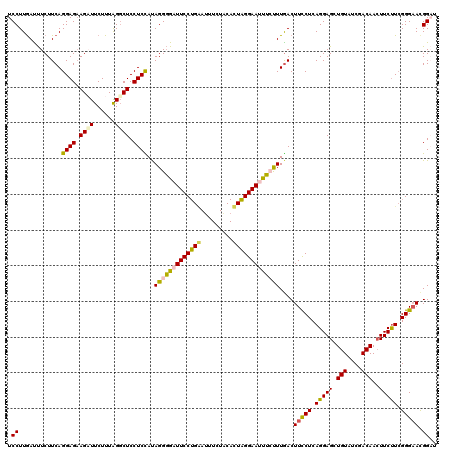
| Location | 3,281,640 – 3,281,759 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.53 |
| Mean single sequence MFE | -35.11 |
| Consensus MFE | -28.25 |
| Energy contribution | -28.64 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3281640 119 - 23771897 GUCUCUGGAGCAGUCUCUUUGAUUUCCUCGGGAGUGCUUUCCUUGAUUUCUUCAGGAGAAGAUUCUUUAGGCUCCUCCAUAGGGGAUUCCUGAAUCUCUACAUUAGGAAUUUGUUUGAC (((...(((((((((.....))))..((..(((((.(((..(((((.....)))))..))))))))..))))))).....((.((((((((((.........)))))))))).)).))) ( -34.80) >DroSec_CAF1 11614 119 - 1 GUCUCUGGAGCAGUCUCUUUGAUUUCCUCGGGAGUGCUUUCCUUGACUUCUUCAGGAGAAGAUUCUUUAGUCUCCUCCAUAGGGGAUUCCUGGACUUCUACACUAGGAAUCUCUUUGAC (((...((((((.((((............)))).)))..)))..)))....(((((((.((((......)))).))))..(((((((((((((.........)))))))))))))))). ( -40.30) >DroSim_CAF1 12465 119 - 1 GUCUCUGGAGCAGUCUCUUUGAUUUCCUCGGGAGUGCUUUCCUUGACUUCUUCAGGAGAAGAUUCUUUAGGCUCCUCCAUAGGGGUUUCCUGAAUUUCUACACUGGGAAUCUCUUUGAC ......(((((((((.....))))..((..(((((.(((..(((((.....)))))..))))))))..)))))))..((.((((((((((..............)))))))))).)).. ( -36.84) >DroEre_CAF1 11813 100 - 1 ----CUGAAGCAGCCUCUUUGAUUUCCUCGGGAGUGGUCUCCUUGAUCUCUUCAGGAGAAGUUUCUUUGGGCUCCUCUACAGGUGAUUCC---------------GGAACUGCCUUAAC ----.(((.((((..(((..((((.(((..((((..((((....((...((((....))))..))...))))..))))..))).))))..---------------))).)))).))).. ( -27.70) >DroYak_CAF1 12250 119 - 1 GACUCUGGAGCAGUCUCUUUGAUUUCCUCGGGAGUGCUUUCCUUAAUUUCUUCAGGAGCAGUUUCUUUGGGCUCCUCUACAGGGGAUUCCUGGAUUUCUGCACUAGGAAUUUGUUUGAC .(((((.(((.((((.....))))..))).)))))........(((.(((((.((..((((..(((..((..(((((....)))))..)).)))...)))).))))))).)))...... ( -35.90) >consensus GUCUCUGGAGCAGUCUCUUUGAUUUCCUCGGGAGUGCUUUCCUUGAUUUCUUCAGGAGAAGAUUCUUUAGGCUCCUCCAUAGGGGAUUCCUGAAUUUCUACACUAGGAAUUUCUUUGAC .....(((((.(((((....(((((.((((((((....)))))(((.....))).))).)))))....))))).))))).(((((((((((((.........))))))))))))).... (-28.25 = -28.64 + 0.39)

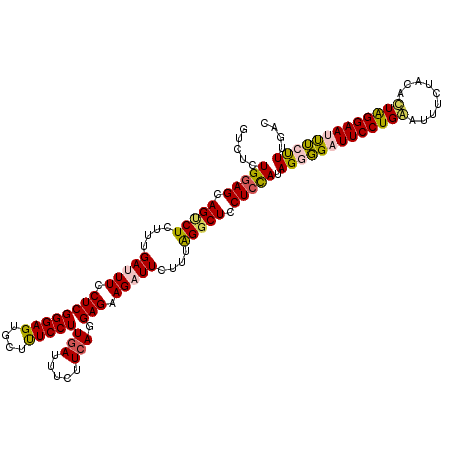
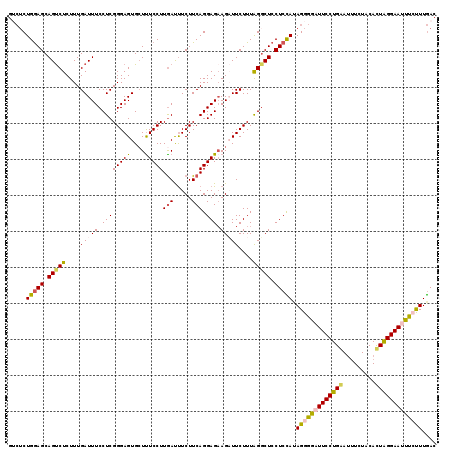
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:36 2006