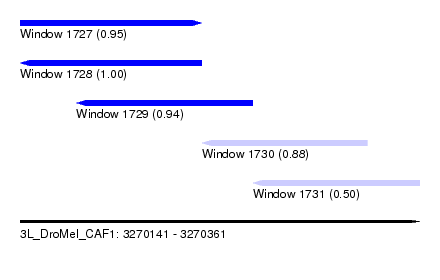
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,270,141 – 3,270,361 |
| Length | 220 |
| Max. P | 0.996401 |

| Location | 3,270,141 – 3,270,241 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 74.00 |
| Mean single sequence MFE | -19.25 |
| Consensus MFE | -7.30 |
| Energy contribution | -7.80 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.38 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
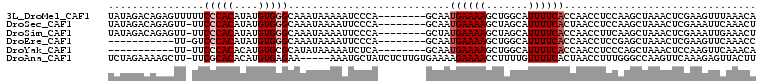
>3L_DroMel_CAF1 3270141 100 + 23771897 UAUAGACAGAGUUUUUCCCACAUAUGUGGGCAAAUAAAAAUCCCA--------GCAAUGAAAAGCUGGCAUUUUCACCAACCUCCAAGCUAAACUCGAAGUUUAAACA ..(((((.((((((..(((((....)))))......(((((.(((--------((........))))).)))))................))))))...))))).... ( -25.10) >DroSec_CAF1 294 99 + 1 UAUAGACAGAGUU-UUCCCACAUAUGUGGGCAAAUAAAAUUCCCA--------GCAAUGAAAAGCUAGCAUUUUCACUAACCUCCAAGCUAAACUCGAAAUUCAAACU ........(((((-(.(((((....)))))...............--------....((((((.......))))))..............))))))............ ( -15.90) >DroSim_CAF1 319 99 + 1 UAUAGACAGAGUU-UUCCCACAUAUGUGGGCAAAUAAAAUUCCCA--------GCUAUGAAAAGCUAGCAUUUUCACCAACCUUCAAGCUAAACUCGAAAUUGAAACU ........(((((-(.(((((....)))))..............(--------(((.((((.....................))))))))))))))............ ( -17.00) >DroEre_CAF1 321 88 + 1 -----------UU-GUCCCACAUAUGUGGGCAAAUAAAAUUCCCA--------GCAAUGAAAAGCUGGCAUUUUCACCAACCUCCGAGCUAAACUCGAAGUUCAAACC -----------((-(.(((((....))))))))..(((((..(((--------((........))))).)))))...........(((((........)))))..... ( -22.20) >DroYak_CAF1 364 88 + 1 -----------UU-UUCCCACACAUGUGCGCAUAUAAAAAUCUCA--------GCAAUGAAAAGCUGGCAUUUUCACCAACCUCCCAGCUAAACUCCAAGUUCAAACA -----------..-..........((.((.............(((--------....)))..((((((................)))))).........)).)).... ( -10.49) >DroAna_CAF1 234 102 + 1 UCUAGAAAAGCUU-UUCGCACACAUGUGAGAA-----AAAUGCUAUCUCUUGUGAAAAGAAAACCUUUUGUUUUCACUAACCUUUGGGCCAAGUUCAAAGAGUUACUU ...(((..(((((-(((.(((....))).)))-----))..)))...))).(((((((.(((....))).)))))))((((((((((((...)))))))).))))... ( -24.80) >consensus UAUAGACAGAGUU_UUCCCACAUAUGUGGGCAAAUAAAAAUCCCA________GCAAUGAAAAGCUGGCAUUUUCACCAACCUCCAAGCUAAACUCGAAGUUCAAACU ................(((((....)))))...........................((((((.......))))))................................ ( -7.30 = -7.80 + 0.50)
| Location | 3,270,141 – 3,270,241 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 74.00 |
| Mean single sequence MFE | -27.87 |
| Consensus MFE | -16.66 |
| Energy contribution | -16.92 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.996401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
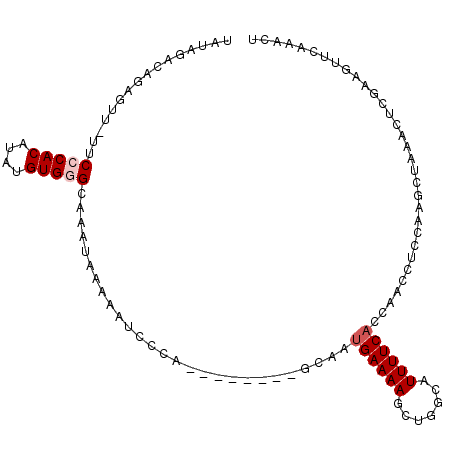
>3L_DroMel_CAF1 3270141 100 - 23771897 UGUUUAAACUUCGAGUUUAGCUUGGAGGUUGGUGAAAAUGCCAGCUUUUCAUUGC--------UGGGAUUUUUAUUUGCCCACAUAUGUGGGAAAAACUCUGUCUAUA .((((...(((((((.....)))))))...(((((((((.(((((........))--------))).)))))))))..(((((....)))))..)))).......... ( -33.30) >DroSec_CAF1 294 99 - 1 AGUUUGAAUUUCGAGUUUAGCUUGGAGGUUAGUGAAAAUGCUAGCUUUUCAUUGC--------UGGGAAUUUUAUUUGCCCACAUAUGUGGGAA-AACUCUGUCUAUA ((((((((((((.(((.......((((((((((......))))))))))....))--------).)))))))......(((((....))))).)-))))......... ( -26.40) >DroSim_CAF1 319 99 - 1 AGUUUCAAUUUCGAGUUUAGCUUGAAGGUUGGUGAAAAUGCUAGCUUUUCAUAGC--------UGGGAAUUUUAUUUGCCCACAUAUGUGGGAA-AACUCUGUCUAUA ............((((((((((.((((((((((......))))))))))...)))--------)..............(((((....))))).)-)))))........ ( -27.80) >DroEre_CAF1 321 88 - 1 GGUUUGAACUUCGAGUUUAGCUCGGAGGUUGGUGAAAAUGCCAGCUUUUCAUUGC--------UGGGAAUUUUAUUUGCCCACAUAUGUGGGAC-AA----------- ........(((((((.....)))))))...(((((((.(.(((((........))--------))).).)))))))(((((((....))))).)-).----------- ( -31.90) >DroYak_CAF1 364 88 - 1 UGUUUGAACUUGGAGUUUAGCUGGGAGGUUGGUGAAAAUGCCAGCUUUUCAUUGC--------UGAGAUUUUUAUAUGCGCACAUGUGUGGGAA-AA----------- ...............((((((((..((((((((......))))))))..))..))--------)))).(((((...(((((....))))).)))-))----------- ( -24.40) >DroAna_CAF1 234 102 - 1 AAGUAACUCUUUGAACUUGGCCCAAAGGUUAGUGAAAACAAAAGGUUUUCUUUUCACAAGAGAUAGCAUUU-----UUCUCACAUGUGUGCGAA-AAGCUUUUCUAGA ...((((.(((((.........)))))))))(((((((.(((....))).))))))).(((((.(((..((-----(((.(((....))).)))-))))))))))... ( -23.40) >consensus AGUUUGAACUUCGAGUUUAGCUUGGAGGUUGGUGAAAAUGCCAGCUUUUCAUUGC________UGGGAAUUUUAUUUGCCCACAUAUGUGGGAA_AACUCUGUCUAUA ........(((((((.....)))))))((((((......)))))).................................(((((....)))))................ (-16.66 = -16.92 + 0.25)
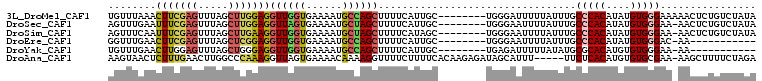
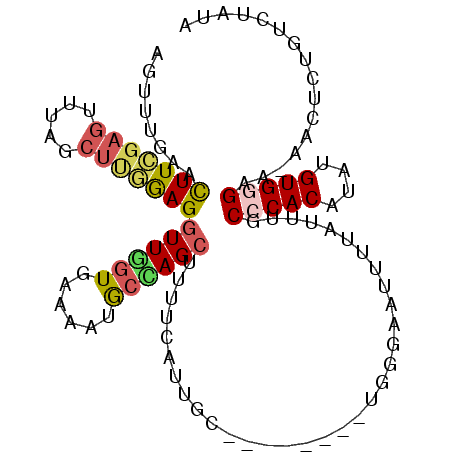
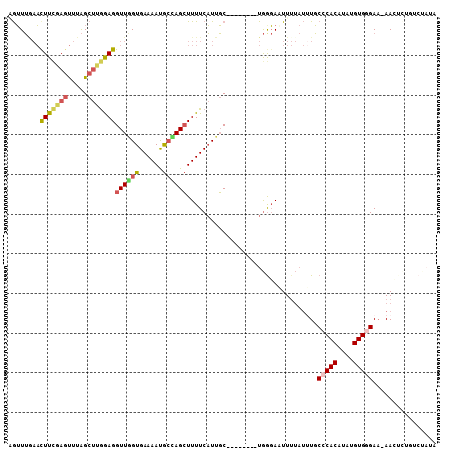
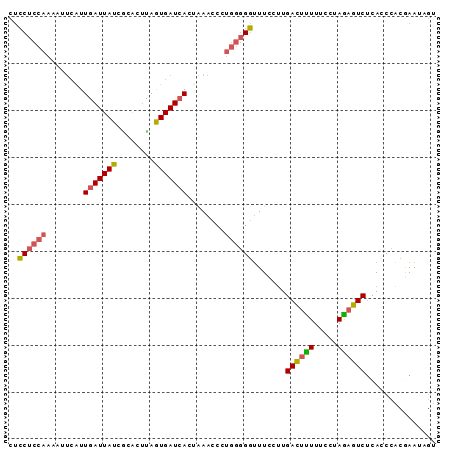
| Location | 3,270,172 – 3,270,269 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 77.19 |
| Mean single sequence MFE | -25.35 |
| Consensus MFE | -14.62 |
| Energy contribution | -14.32 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3270172 97 - 23771897 UUUUCCUAGAGUCUCACCCACGAAUAGUUGUUUAAACUUCGAGUUUAGCUUGGAGGUUGGUGAAAAUGCCAGCUUUUCAUUGC--------UGGGAUUUUUAUUU .......(((((((((((.((((((....))))...(((((((.....))))))))).))))).....(((((........))--------)))))))))..... ( -27.20) >DroSec_CAF1 324 97 - 1 UUUUCCUAGAGUCUCAGCCACGAAUUGUAGUUUGAAUUUCGAGUUUAGCUUGGAGGUUAGUGAAAAUGCUAGCUUUUCAUUGC--------UGGGAAUUUUAUUU ..(((((((......((((.(((((....)))))...((((((.....))))))))))((((((((.(....))))))))).)--------))))))........ ( -24.00) >DroSim_CAF1 349 97 - 1 UUUUCCUAGAGUCUCACCCACGAAUUGUAGUUUCAAUUUCGAGUUUAGCUUGAAGGUUGGUGAAAAUGCUAGCUUUUCAUAGC--------UGGGAAUUUUAUUU ..((((((.....(((((.(((((.......)))..(((((((.....))))))))).)))))....((((........))))--------))))))........ ( -24.30) >DroEre_CAF1 340 97 - 1 CUUUCCUAGAGUCUCACCCACGAAUAGUGGUUUGAACUUCGAGUUUAGCUCGGAGGUUGGUGAAAAUGCCAGCUUUUCAUUGC--------UGGGAAUUUUAUUU ..(((((((........((((.....))))..(((((((((((.....)))))))((((((......))))))..))))...)--------))))))........ ( -32.90) >DroYak_CAF1 383 97 - 1 UUUUCUUAGAGUCUCACCCACAAAUAGUUGUUUGAACUUGGAGUUUAGCUGGGAGGUUGGUGAAAAUGCCAGCUUUUCAUUGC--------UGAGAUUUUUAUAU .......((((((((((((((((((....)))))....))).))..(((((..((((((((......))))))))..))..))--------)))))))))..... ( -28.00) >DroAna_CAF1 264 100 - 1 UUUGGCCAAUUUCCCACCCUCAAGUAACAAGUAACUCUUUGAACUUGGCCCAAAGGUUAGUGAAAACAAAAGGUUUUCUUUUCACAAGAGAUAGCAUUU-----U ..(((........)))..................(((((((((.((((((....)))))).((((((.....))))))..)))).))))).........-----. ( -15.70) >consensus UUUUCCUAGAGUCUCACCCACGAAUAGUAGUUUGAACUUCGAGUUUAGCUUGGAGGUUGGUGAAAAUGCCAGCUUUUCAUUGC________UGGGAAUUUUAUUU ...(((((............................(((((((.....)))))))((((((......))))))..................)))))......... (-14.62 = -14.32 + -0.30)

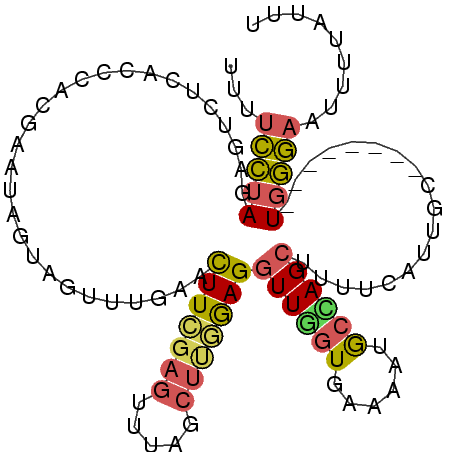
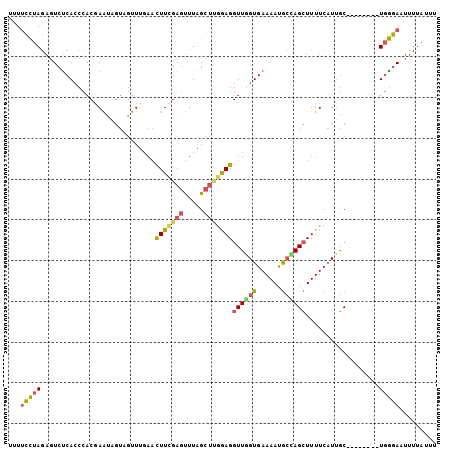
| Location | 3,270,241 – 3,270,332 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 79.69 |
| Mean single sequence MFE | -17.36 |
| Consensus MFE | -14.51 |
| Energy contribution | -14.46 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3270241 91 - 23771897 CUCCUCCAAAAUUCAUUGAUUAUCGCACCUAAUGAUCACUAAACCCUGGGGGUUUCCUUGACUUUUUCCUAGAGUCUCACCCACGAAUAGU ..((((((........(((((((........)))))))........)))))).......((((((.....))))))............... ( -17.19) >DroSec_CAF1 393 91 - 1 CUCCUCCAAAAUUCAUUGAUUAUCGUGCUUAGUGAUCACUAAACCCUGGGGGUUUCCUUGACUUUUUCCUAGAGUCUCAGCCACGAAUUGU ..((((((........(((((((........)))))))........)))))).......((((((.....))))))............... ( -17.49) >DroSim_CAF1 418 91 - 1 CUCCUCCAAAAUUCAUUGAUUAUCGUACUUAGUGAUCACUAAACCCUGGGGGUUUCCUUGACUUUUUCCUAGAGUCUCACCCACGAAUUGU ..((((((........(((((((........)))))))........)))))).......((((((.....))))))............... ( -17.49) >DroEre_CAF1 409 90 - 1 CUCCUCCAAA-GUCAUUGAUUAUCCCACUUAGUGAUCACAAAACCCUGGGGGUUUCCUUGACUCUUUCCUAGAGUCUCACCCACGAAUAGU ..((((((..-((...(((((((........)))))))....))..)))))).......((((((.....))))))............... ( -20.80) >DroYak_CAF1 452 90 - 1 CUCCUCCAAA-CUCAUUGAUUAUCACACUUAGUGAUCACAAAACCCUGGGGGUUUCCUUGACUUUUUCUUAGAGUCUCACCCACAAAUAGU ..((((((..-....(((...(((((.....)))))..))).....)))))).......((((((.....))))))............... ( -17.20) >DroAna_CAF1 336 84 - 1 CCGCCCCGUAGGCGGUUUAUUAUCUCAUUAAAUGAUCACCCAA-------GUGUUCAAUGAAUUUUGGCCAAUUUCCCACCCUCAAGUAAC (((((.....))))).(((((...........(((.(((....-------))).))).(((....(((........)))...)))))))). ( -14.00) >consensus CUCCUCCAAAAUUCAUUGAUUAUCGCACUUAGUGAUCACUAAACCCUGGGGGUUUCCUUGACUUUUUCCUAGAGUCUCACCCACGAAUAGU ..((((((........(((((((........)))))))........)))))).......((((((.....))))))............... (-14.51 = -14.46 + -0.05)

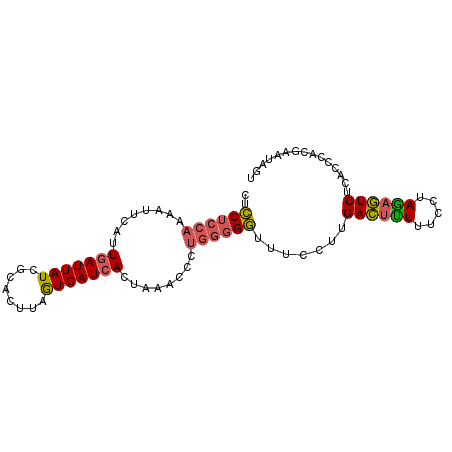
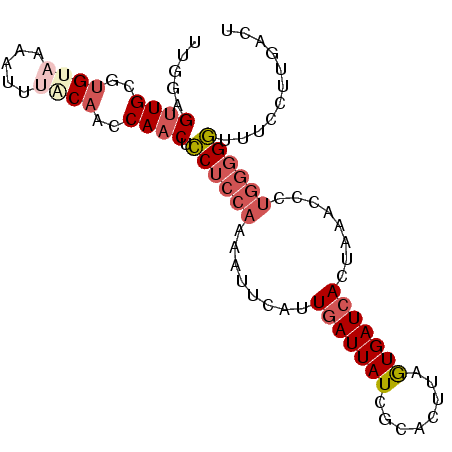
| Location | 3,270,269 – 3,270,361 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 80.57 |
| Mean single sequence MFE | -21.68 |
| Consensus MFE | -11.62 |
| Energy contribution | -12.87 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.54 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3270269 92 - 23771897 UUGGAGUUGCGUGUAAAAUUUACAACCAACUCCUCCAAAAUUCAUUGAUUAUCGCACCUAAUGAUCACUAAACCCUGGGGGUUUCCUUGACU ..(((((((..((((.....))))..)))))))........(((.(((((((........)))))))..((((((...))))))...))).. ( -21.80) >DroSec_CAF1 421 91 - 1 UU-GAGUUGCGUGUAUAAUUUACAACCAACUCCUCCAAAAUUCAUUGAUUAUCGUGCUUAGUGAUCACUAAACCCUGGGGGUUUCCUUGACU ..-((((((..((((.....))))..))))))(((((........(((((((........)))))))........)))))............ ( -20.09) >DroSim_CAF1 446 92 - 1 UUGGAGUUGCGUGUAUAAUUUACAACCAACUCCUCCAAAAUUCAUUGAUUAUCGUACUUAGUGAUCACUAAACCCUGGGGGUUUCCUUGACU ..(((((((..((((.....))))..)))))))........(((.(((((((........)))))))..((((((...))))))...))).. ( -22.90) >DroEre_CAF1 437 91 - 1 UGGGUGUUGCGUGUAAAAUUCCCAAGCAACUCCUCCAAA-GUCAUUGAUUAUCCCACUUAGUGAUCACAAAACCCUGGGGGUUUCCUUGACU .(((.(((((.((.........)).))))).)))....(-((((.(((((((........)))))))..((((((...))))))...))))) ( -22.90) >DroYak_CAF1 480 91 - 1 UUUGUGUUGCGUGUAAAAUUUGCAACCAACUCCUCCAAA-CUCAUUGAUUAUCACACUUAGUGAUCACAAAACCCUGGGGGUUUCCUUGACU .(((.((((((.........)))))))))..((((((..-....(((...(((((.....)))))..))).....))))))........... ( -19.60) >DroAna_CAF1 364 85 - 1 GGGGAGUUGCGUGUAAAAUUCUCCAACAUCCGCCCCGUAGGCGGUUUAUUAUCUCAUUAAAUGAUCACCCAA-------GUGUUCAAUGAAU ((((((((........)))))))).....(((((.....))))).........((((....(((.(((....-------))).))))))).. ( -22.80) >consensus UUGGAGUUGCGUGUAAAAUUUACAACCAACUCCUCCAAAAUUCAUUGAUUAUCGCACUUAGUGAUCACUAAACCCUGGGGGUUUCCUUGACU .....((((..((((.....))))..)))).((((((........(((((((........)))))))........))))))........... (-11.62 = -12.87 + 1.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:27 2006