| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,227,531 – 3,227,645 |
| Length | 114 |
| Max. P | 0.851346 |

| Location | 3,227,531 – 3,227,645 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.15 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -7.37 |
| Energy contribution | -7.98 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.53 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.22 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
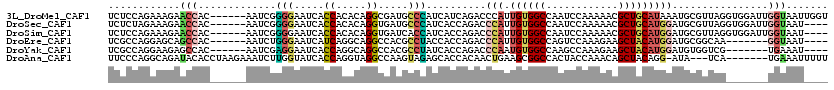
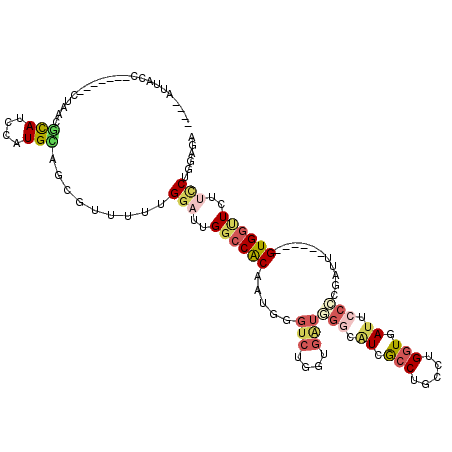
>3L_DroMel_CAF1 3227531 114 + 23771897 UCUCCAGAAAGAACCAC------AAUCGGGGAAUCACCACACAGGCGAUGCCCAUCAUCAGACCCAUUGUGGCCAAUCCAAAAACGCUGCAUAAAUGCGUUAGGUGGAUUGGUAAUUGGU ...((((......((((------(((.((((.(((.((.....)).))).))).((....)).).)))))))((((((((..(((((.........)))))...))))))))...)))). ( -38.00) >DroSec_CAF1 6840 110 + 1 UCUCUAGAAAGAACCAC------AAUCGGGGAAUCACCACACAGGUGAUGCCCAUCACCAGACCCAUUGUGGCCAAUCCAAAAACGCUGCAUGGAUGCGUUAGGUGGAUUGGUAAU---- .............((((------(((.((((.((((((.....)))))).))).((....)).).)))))))((((((((..(((((.........)))))...))))))))....---- ( -39.20) >DroSim_CAF1 6931 110 + 1 UCUCCAGAAAGAACCAC------AAUCGGGGAAUCACCACACAGGUGAUCACCAUCACCAGACCCAUUGUGGCCAAUCCAAAAACGCUGCAUGGAUGCGUUAGGUGGAUUGGUAAU---- .............((((------(((.(((((.(((((.....)))))))....((....))))))))))))((((((((..(((((.........)))))...))))))))....---- ( -38.50) >DroEre_CAF1 6712 103 + 1 UCGCCAGGAGCAGCCAC------AAUCUGGGAAUCAUCAGGCAGGCCACGCCUACCACCAGACCCAUUGUGGCCAGUCCAAAGAAGCUACAUGGAUGCGGCAA-------GGUAAU---- ..(((.(((...(((((------(((((((((....)).((.((((...)))).)).)))))....)))))))...)))......((..((....))..))..-------)))...---- ( -34.70) >DroYak_CAF1 6864 103 + 1 UCGCCAGGAAGAGCCAC------AAUCGAGGAAUCACCAGGCAGGCCACGCCUAUCACCAGACCCAAUGUGGCCAAGCCAAAGAAGCUACAUGGAUGUGGUCG-------UGAAAU---- ..(((.((.....((..------......)).....)).)))((((...)))).((((..(((((((((((((............)))))))...)).)))))-------)))...---- ( -28.30) >DroAna_CAF1 8015 109 + 1 UUCCCAGGCAGAUACACCUAAGAAAUCUUGGUAUCACCAGGUAGGCCAAGUAGAGCACCACAACUGAAGCGGCCACUACCAAACAGCUACAGG-AUA---UCA-------UGAAAUUUUU .(((..(((.(((((...((((....)))))))))....((((((((...(((..........)))....))))..)))).....)))...))-)..---...-------.......... ( -22.30) >consensus UCUCCAGAAAGAACCAC______AAUCGGGGAAUCACCACACAGGCCAUGCCCAUCACCAGACCCAUUGUGGCCAAUCCAAAAAAGCUACAUGGAUGCGUUAG_______GGUAAU____ ............(((.............(((.....((.....)).....)))..........(((.((((((............)))))))))................)))....... ( -7.37 = -7.98 + 0.61)
| Location | 3,227,531 – 3,227,645 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.15 |
| Mean single sequence MFE | -38.28 |
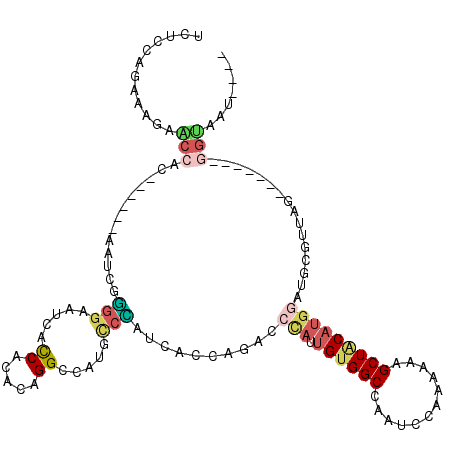
| Consensus MFE | -12.00 |
| Energy contribution | -13.12 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.31 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3227531 114 - 23771897 ACCAAUUACCAAUCCACCUAACGCAUUUAUGCAGCGUUUUUGGAUUGGCCACAAUGGGUCUGAUGAUGGGCAUCGCCUGUGUGGUGAUUCCCCGAUU------GUGGUUCUUUCUGGAGA .(((....((((((((...(((((.........)))))..))))))))(((((((.((((....)))(((.((((((.....)))))).)))).)))------)))).......)))... ( -44.60) >DroSec_CAF1 6840 110 - 1 ----AUUACCAAUCCACCUAACGCAUCCAUGCAGCGUUUUUGGAUUGGCCACAAUGGGUCUGGUGAUGGGCAUCACCUGUGUGGUGAUUCCCCGAUU------GUGGUUCUUUCUAGAGA ----....((((((((...(((((.........)))))..))))))))(((((((.((((....)))(((.((((((.....)))))).)))).)))------))))((((....)))). ( -43.00) >DroSim_CAF1 6931 110 - 1 ----AUUACCAAUCCACCUAACGCAUCCAUGCAGCGUUUUUGGAUUGGCCACAAUGGGUCUGGUGAUGGUGAUCACCUGUGUGGUGAUUCCCCGAUU------GUGGUUCUUUCUGGAGA ----....((((((((...(((((.........)))))..))))))))(((((((.((((....)))((..((((((.....))))))..))).)))------))))((((....)))). ( -41.70) >DroEre_CAF1 6712 103 - 1 ----AUUACC-------UUGCCGCAUCCAUGUAGCUUCUUUGGACUGGCCACAAUGGGUCUGGUGGUAGGCGUGGCCUGCCUGAUGAUUCCCAGAUU------GUGGCUGCUCCUGGCGA ----......-------(((((((.........))......(((..((((((((((((((....(((((((...)))))))....))..))))..))------))))))..))).))))) ( -40.20) >DroYak_CAF1 6864 103 - 1 ----AUUUCA-------CGACCACAUCCAUGUAGCUUCUUUGGCUUGGCCACAUUGGGUCUGGUGAUAGGCGUGGCCUGCCUGGUGAUUCCUCGAUU------GUGGCUCUUCCUGGCGA ----......-------((.(((.........((((.....)))).((((((((((((.(..(.(.(((((...)))))))..).....)).))).)------)))))).....))))). ( -28.80) >DroAna_CAF1 8015 109 - 1 AAAAAUUUCA-------UGA---UAU-CCUGUAGCUGUUUGGUAGUGGCCGCUUCAGUUGUGGUGCUCUACUUGGCCUACCUGGUGAUACCAAGAUUUCUUAGGUGUAUCUGCCUGGGAA .......((.-------((.---(((-(((((((..((..(((((.((((((.......))))).).)))))..))))))..)).)))).)).)).(((((((((......))))))))) ( -31.40) >consensus ____AUUACC_______CUAACGCAUCCAUGCAGCGUUUUUGGAUUGGCCACAAUGGGUCUGGUGAUGGGCAUCGCCUGCCUGGUGAUUCCCCGAUU______GUGGUUCUUCCUGGAGA ......................(((....))).........(((..((((((.....(((....)))(((.((.(((.....))).)).)))...........))))))..)))...... (-12.00 = -13.12 + 1.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:05 2006