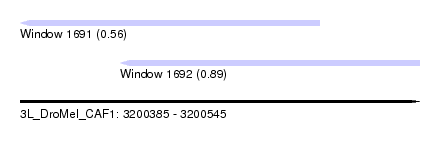
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,200,385 – 3,200,545 |
| Length | 160 |
| Max. P | 0.892410 |

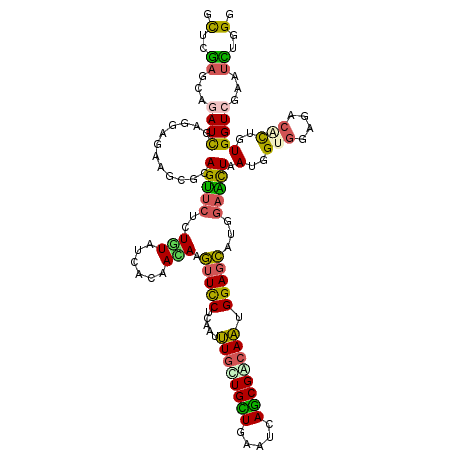
| Location | 3,200,385 – 3,200,505 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.50 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -19.78 |
| Energy contribution | -18.98 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3200385 120 - 23771897 AACAAGUUCCUCAAUUUGUUGUUGAAUCAACGACAAUGGAGCAUGGAACUCAUGGUGGAAACUCUUUGGUCAAAUCUAGGUAACUCCAUCAACGACACCAACAACUUGAAAAAGACUCAU ....((((((....((..((((((......))))))..))....))))))..(((((((....(((.(((...))).)))....))))))).............(((....)))...... ( -28.30) >DroVir_CAF1 20898 120 - 1 AAUAAGUUCCUCAAUUUACUGCUCAAUCAGCGGCAAUGGAGCAUGGAAUUGAUGGUGGAGACGCUCUGGUCUAAUCUGGGCAACUCCAACAACGACACAAACAAUCUGAAAAAGACGCAU .....((((((((((((..(((((.((........)).)))))..))))))).))(((((..((((.(((...))).))))..))))).....)))........(((.....)))..... ( -27.60) >DroGri_CAF1 22603 120 - 1 AACAAGUUUCUCAAUUUGCUGCUGAAUAAGCGGCAAUGGAGCAUGGAAUUAAUGGUGGAGACAUUGUGGUCCAAUUUGGGUAACUCCAAUAACGACACCAAUAACCUCAAGAAGACACAU .....((((.((...((((((((.....))))))))(((....((((....(..(((....)))..)..))))..(((((....)))))........)))..........)))))).... ( -32.10) >DroWil_CAF1 19556 120 - 1 AAUAAAUUUCUCAAUUUGCUGCUAAAUCAGCGACAAUGGACUAUGGAGCUAAUGGUAGAGACUCUUUGGUCGAAUUUGGGCAAUUCUAUAAAUGAUACAAACAAUCUAAAGAAGACCCAU .......(((.....((((((......)))))).....(((((.((((((........)).)))).))))))))..((((...((((......(((.......)))...))))..)))). ( -23.80) >DroMoj_CAF1 19019 120 - 1 AAUAAGUUUCUUAAUUUGUUGCUCAAUCAGCGGCAAUGGAGCAUGGAGCUAAUGGUGGAAACGCUGUGGUCUAAUCUGGGCAACUCCAACAACGACACAAACAAUCUAAAGAAGACGCAU .....(((((.....((((((((.....)))))))).))))).(((((...((((((....)))))).((((.....))))..)))))................................ ( -28.80) >DroAna_CAF1 18749 120 - 1 AACAAGUUCCUGAAUCUGCUGUUGGGCCAGCGACAGUGGAGCAUGGAGCUCAUGGUGGAGACACUGUGGUCGAACUUGGGCAAUUCCAUAAACGAUACCAACAACCUGAAGAAGACCCAU ....((((((....((..((((((......))))))..))....))))))(((((((....)))))))((((....((((....))))....))))........................ ( -38.30) >consensus AACAAGUUCCUCAAUUUGCUGCUGAAUCAGCGACAAUGGAGCAUGGAACUAAUGGUGGAGACACUGUGGUCGAAUCUGGGCAACUCCAACAACGACACAAACAACCUGAAGAAGACCCAU .....(((((.....((((((((.....)))))))).))))).((((....(..(((....)))..).(((.......)))...))))................................ (-19.78 = -18.98 + -0.80)

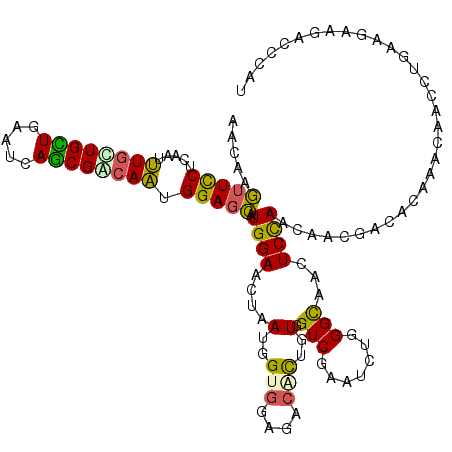
| Location | 3,200,425 – 3,200,545 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.67 |
| Mean single sequence MFE | -37.90 |
| Consensus MFE | -20.65 |
| Energy contribution | -19.38 |
| Covariance contribution | -1.27 |
| Combinations/Pair | 1.56 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3200425 120 - 23771897 CUUGGAGCAGAUCGAGGAAAAGCGAAGUAAUCUCUAUGAGAACAAGUUCCUCAAUUUGUUGUUGAAUCAACGACAAUGGAGCAUGGAACUCAUGGUGGAAACUCUUUGGUCAAAUCUAGG ((((((...(((((((((...............(((((((..((.(((((.....(((((((((...))))))))).))))).))...))))))).(....))))))))))...)))))) ( -40.70) >DroVir_CAF1 20938 120 - 1 ACUGGAGCAAAUUGAGGAGAAGCGGAGUUCUCUGUAUCGCAAUAAGUUCCUCAAUUUACUGCUCAAUCAGCGGCAAUGGAGCAUGGAAUUGAUGGUGGAGACGCUCUGGUCUAAUCUGGG .(..((((((((((((((((.(((((....))))).))((.....)))))))))))).(((((.....))))).......)).((((......((((....))))....)))).))..). ( -36.90) >DroGri_CAF1 22643 120 - 1 GCUCGAACAGAUUGAGGAGAAACGCAGUUCGCUGUAUCGCAACAAGUUUCUCAAUUUGCUGCUGAAUAAGCGGCAAUGGAGCAUGGAAUUAAUGGUGGAGACAUUGUGGUCCAAUUUGGG ((((............(((((((...(((.((......)))))..)))))))...((((((((.....))))))))..)))).((((....(..(((....)))..)..))))....... ( -42.40) >DroWil_CAF1 19596 120 - 1 UCUCGAACAGAUUGAGGAGAAGCGCAGCUCACUGUAUAAUAAUAAAUUUCUCAAUUUGCUGCUAAAUCAGCGACAAUGGACUAUGGAGCUAAUGGUAGAGACUCUUUGGUCGAAUUUGGG .((((((..(((..(((((....((((....)))).............((((...((((((......))))))......(((((.......))))).)))))))))..)))...)))))) ( -31.50) >DroMoj_CAF1 19059 120 - 1 GCUCGAGCAGAUCGAGGAAAAACGUAGUUCUCUAUAUCGCAAUAAGUUUCUUAAUUUGUUGCUCAAUCAGCGGCAAUGGAGCAUGGAGCUAAUGGUGGAAACGCUGUGGUCUAAUCUGGG .((.((..(((((...........(((((((.......((((((((((....))))))))))..........((......))..)))))))((((((....)))))))))))..)).)). ( -32.50) >DroAna_CAF1 18789 120 - 1 GUUGAAGCAAAUCGAGGAGAAGAGGAGCUCUGUGUAUGAGAACAAGUUCCUGAAUCUGCUGUUGGGCCAGCGACAGUGGAGCAUGGAGCUCAUGGUGGAGACACUGUGGUCGAACUUGGG ...........(((((..((....(((((((((((....(((....))).....((..((((((......))))))..)))))))))))))((((((....))))))..))...))))). ( -43.40) >consensus GCUCGAGCAGAUCGAGGAGAAGCGCAGUUCUCUGUAUCACAACAAGUUCCUCAAUUUGCUGCUGAAUCAGCGACAAUGGAGCAUGGAACUAAUGGUGGAGACACUGUGGUCGAAUCUGGG .(..((...((((............(((((..(((......))).(((((.....((((((((.....)))))))).)))))...))))).(..(((....)))..)))))...))..). (-20.65 = -19.38 + -1.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:51 2006