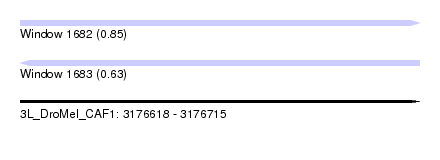
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,176,618 – 3,176,715 |
| Length | 97 |
| Max. P | 0.848511 |

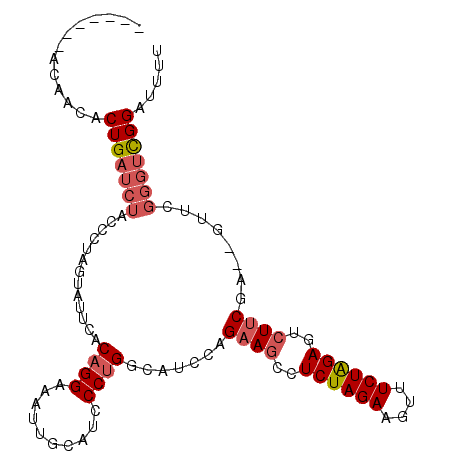
| Location | 3,176,618 – 3,176,715 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 78.53 |
| Mean single sequence MFE | -28.28 |
| Consensus MFE | -17.94 |
| Energy contribution | -18.83 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3176618 97 + 23771897 -------ACAACACUGAUCUACCCUAGUAUUCACAGGAAGUUGCAUCCCUGGCAUCCAGAAGCCUCUAGAAGUUUCUAGAGACUUCCA--GUUCGGGUCGGGUUUU -------..(((.(((((((......((....)).((((((.......((((...))))....(((((((....))))))))))))).--....)))))))))).. ( -28.00) >DroSec_CAF1 13663 97 + 1 -------ACAACACUGAUCUACCCUAGUAUUCACAGGAAGUUGCAUCCCUGGCAACCAGAAGCCUCUAGAAGUUUCUAGAGUCUUCGA--GUUCGGGUCGGUUUUU -------.....((((((((..(((.((....)))))..(((((.......)))))..((((.(((((((....))))))).))))..--....)))))))).... ( -29.60) >DroSim_CAF1 16909 97 + 1 -------ACAACACUGAUCUACCCCAGUAUUCACAGGAAGUUGCAUCCCUGGCAACCAGAAGCCUCUAGAAGUUUCUAGAGUCUUCGA--GUUCGGGUCGGUUUUU -------........((((.((((....((((.......(((((.......)))))..((((.(((((((....))))))).))))))--))..)))).))))... ( -29.60) >DroEre_CAF1 13769 97 + 1 -------ACAACACUGAUCUACGUUAAUAUUCACAGGAAAUUGCAUCCCUGGCAUCUGGAAGACUCUAGAAGUUUCUGGACGCUUCGA--UUUCGGGUCGGAUUUU -------......(((((((..((........))..((((((((.(((..(((.((((((....)))))).)))...))).))...))--)))))))))))..... ( -24.20) >DroYak_CAF1 13923 97 + 1 -------ACAACACUGAUCUCCCUGAGUACGCACAGGAAAUUGCAUCCCUGGCAUCUGGAAGCCUCUAGAAGUUUCUAGACUCUUCGA--GUUCGGGUCGGAUUUU -------........(((((.((((((...((.((((..........))))))..((.((((..((((((....))))))..)))).)--)))))))..))))).. ( -29.50) >DroPer_CAF1 14436 94 + 1 CUCGGUG-CAACUCUG-----UGCCA----GUACCGGAAAUAGCAGCCCUGGAUUCUCGUAGCCUCUAGAAACGUCUAGAAAAUUCUACGCUUGGGGUUGG--UUU .((((((-(.......-----.....----)))))))......((((((..(.....(((((..((((((....)))))).....))))).)..)))))).--... ( -28.80) >consensus _______ACAACACUGAUCUACCCUAGUAUUCACAGGAAAUUGCAUCCCUGGCAUCCAGAAGCCUCUAGAAGUUUCUAGAGUCUUCGA__GUUCGGGUCGGAUUUU .............(((((((.............((((..........)))).......((((..((((((....))))))..))))........)))))))..... (-17.94 = -18.83 + 0.89)

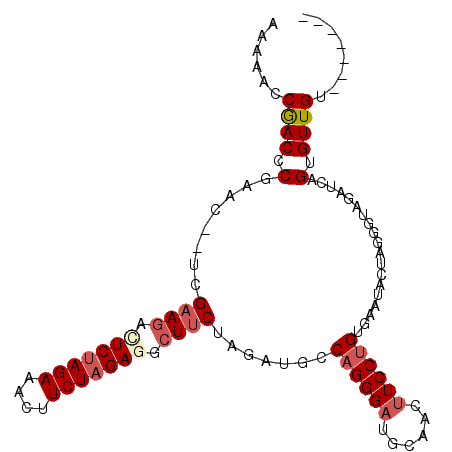
| Location | 3,176,618 – 3,176,715 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 78.53 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -17.49 |
| Energy contribution | -18.43 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3176618 97 - 23771897 AAAACCCGACCCGAAC--UGGAAGUCUCUAGAAACUUCUAGAGGCUUCUGGAUGCCAGGGAUGCAACUUCCUGUGAAUACUAGGGUAGAUCAGUGUUGU------- ........((((...(--..((((((((((((....))))))))))))..)....((((((......)))))).........)))).............------- ( -35.10) >DroSec_CAF1 13663 97 - 1 AAAAACCGACCCGAAC--UCGAAGACUCUAGAAACUUCUAGAGGCUUCUGGUUGCCAGGGAUGCAACUUCCUGUGAAUACUAGGGUAGAUCAGUGUUGU------- ........((((.(((--(.((((.(((((((....))))))).)))).))))..((((((......)))))).........)))).............------- ( -29.60) >DroSim_CAF1 16909 97 - 1 AAAAACCGACCCGAAC--UCGAAGACUCUAGAAACUUCUAGAGGCUUCUGGUUGCCAGGGAUGCAACUUCCUGUGAAUACUGGGGUAGAUCAGUGUUGU------- ......(((((.(...--.)((((.(((((((....))))))).)))).))))).((((((......))))))..((((((((......))))))))..------- ( -30.60) >DroEre_CAF1 13769 97 - 1 AAAAUCCGACCCGAAA--UCGAAGCGUCCAGAAACUUCUAGAGUCUUCCAGAUGCCAGGGAUGCAAUUUCCUGUGAAUAUUAACGUAGAUCAGUGUUGU------- ......((((.(...(--((...(((((..(((((((...)))).)))..)))))((((((......))))))..............)))..).)))).------- ( -19.40) >DroYak_CAF1 13923 97 - 1 AAAAUCCGACCCGAAC--UCGAAGAGUCUAGAAACUUCUAGAGGCUUCCAGAUGCCAGGGAUGCAAUUUCCUGUGCGUACUCAGGGAGAUCAGUGUUGU------- ......(((((.((.(--((((((..((((((....))))))..))))...((((((((((......)))))).)))).......))).)).).)))).------- ( -31.60) >DroPer_CAF1 14436 94 - 1 AAA--CCAACCCCAAGCGUAGAAUUUUCUAGACGUUUCUAGAGGCUACGAGAAUCCAGGGCUGCUAUUUCCGGUAC----UGGCA-----CAGAGUUG-CACCGAG ...--.((((......(((((....(((((((....))))))).))))).....((((..(((.......)))..)----)))..-----....))))-....... ( -22.40) >consensus AAAAACCGACCCGAAC__UCGAAGACUCUAGAAACUUCUAGAGGCUUCUAGAUGCCAGGGAUGCAACUUCCUGUGAAUACUAGGGUAGAUCAGUGUUGU_______ ......((((.(........((((.(((((((....))))))).)))).......((((((......))))))...................).))))........ (-17.49 = -18.43 + 0.95)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:42 2006