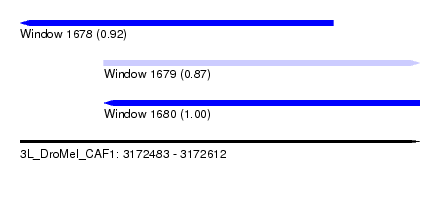
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,172,483 – 3,172,612 |
| Length | 129 |
| Max. P | 0.998814 |

| Location | 3,172,483 – 3,172,584 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.74 |
| Mean single sequence MFE | -20.20 |
| Consensus MFE | -9.05 |
| Energy contribution | -8.50 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
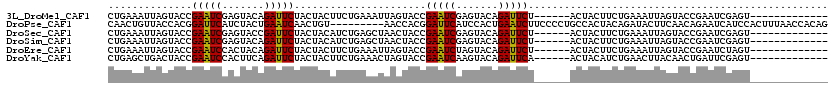
>3L_DroMel_CAF1 3172483 101 - 23771897 CUGAAAUUAGUACCGAAUCGAGUACAGAUUCUACUACUUCUGAAAUUAGUACCGAAUCGAGUACAGAUUCU------ACUACUUCUGAAAUUAGUACCGAAUCGAGU------------- .........(((((((.(((.((((.(((((..........).)))).))))))).))).)))).((((((------((((.((....)).)))))..)))))....------------- ( -22.60) >DroPse_CAF1 11367 111 - 1 CAACUGUUACCACGGAUUCAUCUACUGAAUCAACUGU---------AACCACGGAUUCAUCCACUGAAUCUUCCCCUGCCACUACAGAUACUUCAACAGAAUCAUCCACUUUAACCACAG ...((((.......((((((.....))))))..((((---------......(((((((.....)))))))....(((......)))........)))).................)))) ( -18.30) >DroSec_CAF1 9618 101 - 1 CUGAAAUUAGUACCGAAUCGAGUACCGAUUCUACUACAUCUGAGCUAACUACCGAAUCGAGUACAGAUUCU------ACUACUUCUGAAAUUAGUACCGAAUCGAGU------------- .......(((((..((((((.....)))))))))))................(((.(((.((((.(((((.------.........).)))).))))))).)))...------------- ( -19.70) >DroSim_CAF1 9915 101 - 1 CUGAAAUUAGUACCGAAUCGAGUACAGAUUCUACUACAUCUGAGCUAACUACCGAAUCGAGUACAGAUUCU------ACUACUUCUGAAAUUAGUACCGAAUCGAGU------------- .........(((((((.((((((.(((((........))))).)))......))).))).)))).((((((------((((.((....)).)))))..)))))....------------- ( -21.90) >DroEre_CAF1 9537 101 - 1 CUGAAAUUAGUACCGAAUCCACUACAGAUUCUACUACUUCUGAAAUUAGUACCGAAUCUAGUACAGAUUCU------ACUACUUCUGAAAUUAGUACCGAAUCUAGU------------- .........................(((((((((((.(((.(((..(((((..((((((.....)))))))------)))).))).)))..)))))..))))))...------------- ( -19.00) >DroYak_CAF1 9847 101 - 1 CUGAGCUGACUACCGAAUCCACUUCAGAUUCUACUACUUCUGAAACUAGUACCGAAUCAAGUACAGAUUCA------ACUACAUCUGAACUUACAACUGAUUCGAGU------------- .............((((((.......((((((((((.((....)).)))))..)))))((((.(((((...------.....))))).))))......))))))...------------- ( -19.70) >consensus CUGAAAUUAGUACCGAAUCGACUACAGAUUCUACUACUUCUGAAAUAACUACCGAAUCGAGUACAGAUUCU______ACUACUUCUGAAAUUAGUACCGAAUCGAGU_____________ ..............(((((.......)))))......................(((((.......))))).................................................. ( -9.05 = -8.50 + -0.55)
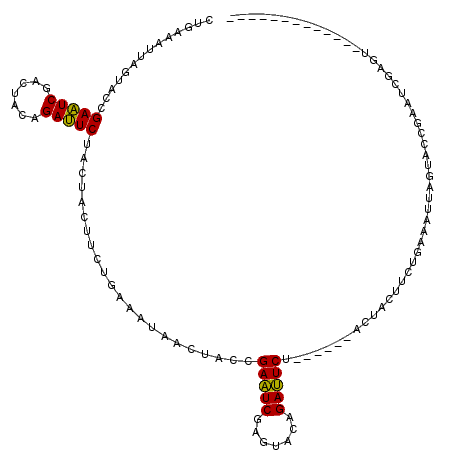
| Location | 3,172,510 – 3,172,612 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.40 |
| Mean single sequence MFE | -29.16 |
| Consensus MFE | -9.71 |
| Energy contribution | -9.22 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3172510 102 + 23771897 AGU------AGAAUCUGUACUCGAUUCGGUACUAAUUUCAGAAGUAGUAGAAUCUGUACUCGAUUCGGUACUAAUUUCAGA---AGUAGUAGAAUCUGUACUCG---------AUUCGGU ...------.(((((.((((..(((((..(((((.((((.((((((((((((((.......)))))..)))).))))).))---)))))))))))).))))..)---------))))... ( -29.20) >DroPse_CAF1 11407 111 + 1 GGCAGGGGAAGAUUCAGUGGAUGAAUCCGUGGUU---------ACAGUUGAUUCAGUAGAUGAAUCCGUGGUAACAGUUGAUUCAGUGGAUGAAUCUGUAGUGGAAGGUGAAGAUUCACU .........(((((((.....((((((....(((---------((....((((((.....))))))....)))))....)))))).....))))))).........(((((....))))) ( -30.60) >DroSec_CAF1 9645 102 + 1 AGU------AGAAUCUGUACUCGAUUCGGUAGUUAGCUCAGAUGUAGUAGAAUCGGUACUCGAUUCGGUACUAAUUUCAGA---UGUAGUAGAAUCGGUACUCG---------AUUCCGU ...------.(((((.((((.((((((.........((.((((.(((((((((((.....))))))..))))))))).)).---.......))))))))))..)---------))))... ( -28.03) >DroSim_CAF1 9942 102 + 1 AGU------AGAAUCUGUACUCGAUUCGGUAGUUAGCUCAGAUGUAGUAGAAUCUGUACUCGAUUCGGUACUAAUUUCAGA---UGUAGUAGAAUCGGUACUCG---------AUUCCGU ...------.(((((.((((.((((((.........((.((((.((((((((((.......)))))..))))))))).)).---.......))))))))))..)---------))))... ( -25.53) >DroEre_CAF1 9564 102 + 1 AGU------AGAAUCUGUACUAGAUUCGGUACUAAUUUCAGAAGUAGUAGAAUCUGUAGUGGAUUCGGUACUAAUUUCAGA---AGUAGUAGAAUCUGUACUGG---------AUUCGGU ...------.((((((((((.((((((..(((((.((((.(((((((((((((((.....))))))..)))).))))).))---))))))))))))))))).))---------))))... ( -34.40) >DroYak_CAF1 9874 102 + 1 AGU------UGAAUCUGUACUUGAUUCGGUACUAGUUUCAGAAGUAGUAGAAUCUGAAGUGGAUUCGGUAGUCAGCUCAGA---UGUAGUAGAAUCUGAAGUGG---------AUUCGGU ..(------((((((((((((......)))))...(((((((..((.((..((((((.((.(((......))).)))))))---).)).))...))))))).))---------)))))). ( -27.20) >consensus AGU______AGAAUCUGUACUCGAUUCGGUACUAAUUUCAGAAGUAGUAGAAUCUGUACUCGAUUCGGUACUAAUUUCAGA___AGUAGUAGAAUCUGUACUCG_________AUUCGGU ..........(((((.......)))))......................((((((.....))))))...................................................... ( -9.71 = -9.22 + -0.50)

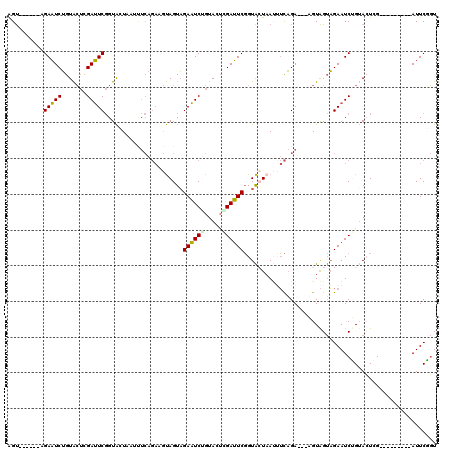
| Location | 3,172,510 – 3,172,612 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.40 |
| Mean single sequence MFE | -26.07 |
| Consensus MFE | -9.05 |
| Energy contribution | -8.50 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.20 |
| Mean z-score | -4.34 |
| Structure conservation index | 0.35 |
| SVM decision value | 3.24 |
| SVM RNA-class probability | 0.998814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3172510 102 - 23771897 ACCGAAU---------CGAGUACAGAUUCUACUACU---UCUGAAAUUAGUACCGAAUCGAGUACAGAUUCUACUACUUCUGAAAUUAGUACCGAAUCGAGUACAGAUUCU------ACU ...((((---------(..((((.((((((((((.(---((.(((..(((((..(((((.......)))))))))).))).)))..)))))..)))))..)))).))))).------... ( -27.90) >DroPse_CAF1 11407 111 - 1 AGUGAAUCUUCACCUUCCACUACAGAUUCAUCCACUGAAUCAACUGUUACCACGGAUUCAUCUACUGAAUCAACUGU---------AACCACGGAUUCAUCCACUGAAUCUUCCCCUGCC .((((....))))..........(((((((.....((((((....(((((....((((((.....))))))....))---------)))....)))))).....)))))))......... ( -26.10) >DroSec_CAF1 9645 102 - 1 ACGGAAU---------CGAGUACCGAUUCUACUACA---UCUGAAAUUAGUACCGAAUCGAGUACCGAUUCUACUACAUCUGAGCUAACUACCGAAUCGAGUACAGAUUCU------ACU ..(((((---------(..((((((((((.......---((.((...(((((..((((((.....)))))))))))..)).))..........)))))).)))).))))))------... ( -28.03) >DroSim_CAF1 9942 102 - 1 ACGGAAU---------CGAGUACCGAUUCUACUACA---UCUGAAAUUAGUACCGAAUCGAGUACAGAUUCUACUACAUCUGAGCUAACUACCGAAUCGAGUACAGAUUCU------ACU ..(((((---------(..(((((((((((((((..---........)))))........(((.(((((........))))).))).......)))))).)))).))))))------... ( -26.70) >DroEre_CAF1 9564 102 - 1 ACCGAAU---------CCAGUACAGAUUCUACUACU---UCUGAAAUUAGUACCGAAUCCACUACAGAUUCUACUACUUCUGAAAUUAGUACCGAAUCUAGUACAGAUUCU------ACU ...((((---------(..(((((((((((((((.(---((.(((..(((((..(((((.......)))))))))).))).)))..)))))..)))))).)))).))))).------... ( -27.00) >DroYak_CAF1 9874 102 - 1 ACCGAAU---------CCACUUCAGAUUCUACUACA---UCUGAGCUGACUACCGAAUCCACUUCAGAUUCUACUACUUCUGAAACUAGUACCGAAUCAAGUACAGAUUCA------ACU ...((((---------(...((((((.........(---((((((.((...........)).))))))).........))))))....((((........)))).))))).------... ( -20.67) >consensus ACCGAAU_________CCAGUACAGAUUCUACUACA___UCUGAAAUUAGUACCGAAUCGACUACAGAUUCUACUACUUCUGAAAUAACUACCGAAUCGAGUACAGAUUCU______ACU ......................................................(((((.......)))))......................(((((.......))))).......... ( -9.05 = -8.50 + -0.55)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:39 2006