
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,158,575 – 3,158,746 |
| Length | 171 |
| Max. P | 0.881087 |

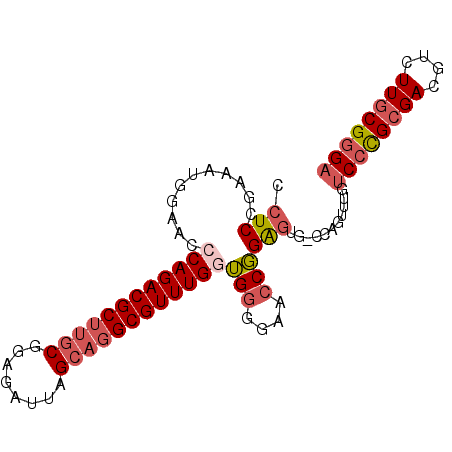
| Location | 3,158,575 – 3,158,666 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 86.71 |
| Mean single sequence MFE | -39.12 |
| Consensus MFE | -29.90 |
| Energy contribution | -31.02 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.637449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3158575 91 + 23771897 CCUCUGAUAUGGAACCCAGACGCUUGCGGAGAUUAGCAGGCGUUUGGUGGGGAACCGGAGUGACCAGUUUGUCCCGCGACGUCUUGCGGGA .((((..........((((((((((((........)))))))))))).((....))))))...........((((((((....)))))))) ( -39.50) >DroSec_CAF1 5558 88 + 1 CCUCCGAUAUGGAACCCAGACGCUUGCGGAGAUUAGCAGGCGUUUGGUGGGGAACCGGAGUG---AGUUUGUCCCGCGACGUCUUGCGGGA .((((..........((((((((((((........)))))))))))).((....))))))..---......((((((((....)))))))) ( -42.20) >DroSim_CAF1 5929 88 + 1 CCUCCGAUAUGGAACCCAGACGCUUGCGGAGAUUAGCAGGCGUUUGGUGGGGAACCGGAGUG---AGUUUGUCCCGCGACGUCUUGCGGGA .((((..........((((((((((((........)))))))))))).((....))))))..---......((((((((....)))))))) ( -42.20) >DroEre_CAF1 6630 91 + 1 CCUCAGAAAUGGAACCCAGACGCUUGCGGAGAUUAGCAGGCGUUUGAUGGGGAACCGGAGUGUCCAGUUUGUCCCGCGAAGCCUUGCGGGA ...((((..((((.(((((((((((((........)))))))))))..((....))))....)))).))))((((((((....)))))))) ( -39.80) >DroYak_CAF1 6197 91 + 1 CCUCAGAAAUGGAACCCAGACGCUUGCGGAGAUUAGCAGGCGUUUGGUGGGGAACCGGAGUGACCAGUUUGUCCCGCGACGUCUUGCGGGC ...((((..(((...((((((((((((........))))))))))))(((....)))......))).)))).(((((((....))))))). ( -38.60) >DroAna_CAF1 5391 88 + 1 CCUCCGAAAUGGAUCCCAGACGCAUUCGCAGAUUGGCCGGCGUUUGAGGAGG---CUGGUGGGCCAGCUUAUCUUGCGAUGUCUUUCGCGA ..(((.....)))((((((((((....((......))..))))))).)))((---((((....))))))....((((((......)))))) ( -32.40) >consensus CCUCCGAAAUGGAACCCAGACGCUUGCGGAGAUUAGCAGGCGUUUGGUGGGGAACCGGAGUG_CCAGUUUGUCCCGCGACGUCUUGCGGGA .(((...........((((((((((((........))))))))))))(((....))))))...........((((((((....)))))))) (-29.90 = -31.02 + 1.11)

| Location | 3,158,643 – 3,158,746 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 71.29 |
| Mean single sequence MFE | -40.47 |
| Consensus MFE | -16.27 |
| Energy contribution | -16.50 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3158643 103 - 23771897 CUGUCAGCUCCACGGAACUCUUGUAUGAAAGGGCCAUGGCCAGGUUCUACGAAGCAGUGGAACUCGAGCAGGCUUCCAAGUCC----CGCAAGACGUC----GCGGGACAA .............((((..(((((.(((...(((....)))..(((((((......)))))))))).))))).))))..((((----(((........----))))))).. ( -40.10) >DroVir_CAF1 6455 109 - 1 CGGUCAGCUCCACGGAGCUACUCUACGAGCGUGCCAUGGCCCGUUUUUACGAGGCCGUGGAACUGGAGCAGGCGGCCAAAAGCAAGGUGA-AGA-CUGCGGCACAGGAGAA .....(((((....))))).((((..(.((((.(((((((((((....))).)))))))).))....((((.(.(((........)))..-.).-)))).)).).)))).. ( -41.70) >DroWil_CAF1 5880 109 - 1 CAGUUAGCUCCACUGAAUUGCUUUAUGAACGGGCCAUGGCCCGAUUCUAUGAGGCUGUGGAGCUGGAACAGGCAUCGAAAUCCGCGCCGAAA--CCUGAAAUACCGGAAAA ...((((((((((......(((((((((((((((....))))).))).))))))).))))))))))..((((..(((..........)))..--))))............. ( -42.50) >DroMoj_CAF1 6091 106 - 1 CGGUCAGCUCCACGGAGCUCCUCUACGAACGUGCCAUGGCGCGUUUCUACGAAGCCGUGGAAUUGGAGCAAGCCUCCAAAUCAAAGGCGA-AGA-CUG---CAAAGGCCCA .(((((((((....)))))..(((.(((((((((....)))))))).......(((...((.((((((.....)))))).))...)))).-)))-...---....)))).. ( -40.50) >DroAna_CAF1 5456 103 - 1 CCGUUAGCUCCACGGAGCUGCUCUACGAGCGUGCGAUGGCCCGGUUCUAUGAGGCCGUAGAACUGGAGCAGGCCUCCAAGUCG----CGAAAGACAUC----GCAAGAUAA .....(((((....)))))((((...)))).((((((((((((((((((((....))))))))))).)).(((......))).----(....).))))----)))...... ( -43.80) >DroPer_CAF1 22245 103 - 1 CCGUGAGCUCCACCGAGCUGCUCUACGAGCGAGCAAUGGCGAGAUUCUAUGAGGCGGUAGAGCUGGAGCAAGCCGCCAAGUCG----AGAAAGGCCUC----CCAGGACAG ((...(((((....)))))((((.......))))..(((.(((.........((((((...((....))..))))))..(((.----.....))))))----))))).... ( -34.20) >consensus CCGUCAGCUCCACGGAGCUGCUCUACGAACGUGCCAUGGCCCGAUUCUACGAGGCCGUGGAACUGGAGCAGGCCUCCAAAUCC____CGAAAGACCUC____ACAGGACAA ..(((.((((((.................((.((....)).)).((((((......)))))).)))))).)))...................................... (-16.27 = -16.50 + 0.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:32 2006