
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 372,263 – 372,355 |
| Length | 92 |
| Max. P | 0.798320 |

| Location | 372,263 – 372,355 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 73.96 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -17.71 |
| Energy contribution | -18.44 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 372263 92 + 23771897 UGCAUUUUGGGUGGGGCCUGGGCAACACCUUUGCGAAUCCC---------------UGGAUUUUCAUCUCCUUAUUGG--AGGGAAUAUAUAUAUUCUGAUGGAAACUA .(((....(((((..((....))..))))).))).......---------------(((.(((((((((((.....))--).((((((....))))))))))))))))) ( -29.40) >DroPse_CAF1 31165 102 + 1 UGCUUUCUGGGUGGAGCCCCGGCAACACCUUUGCGGACGCUGCUCUCCUCCAGAGGUGGAUUUUC---UCCCUCGAGGAGUUGCCUCG----GAGUCUGAUGGAAAUCG .(.((((((((((..((....))..)))))...(((((.((((.((((((..((((..(.....)---..))))))))))..))...)----).)))))..))))).). ( -38.70) >DroSec_CAF1 26359 88 + 1 UGCAUUUUGGGUGGGGCCUGGGCAACACCUUUGCGGAUCCC---------------CGGAUUUUCAUCUCCUUAUUGG--AGGCAUCG----GCUUCUGAUGGAAACUA .(((....(((((..((....))..))))).)))(..(((.---------------.(((........)))..((..(--((((....----)))))..)))))..).. ( -30.40) >DroSim_CAF1 27169 88 + 1 UGCAUUUUGGGUGGGGCCUGGGCAACACCUUUGCGGAUCCC---------------UGGAUUUUCAUCUCCUUAUUGG--AGGGAUCG----GCUUCUGAUGGAAACUA ..((((..(((((..((....))..)))))..((.((((((---------------(((((....))))((.....))--))))))).----))....))))....... ( -30.40) >DroYak_CAF1 28107 88 + 1 UGCAUUUUGGGUGGAGCCUGGGCAACACCUUUGCGGAUCCC---------------UGGAUUUUCAUCUCCCUAUUGG--UGGGAUCG----GCUGCUGAUGGAAACUA .(((....(((((..((....))..)))))..((.((((((---------------.((((....)))).((....))--.)))))).----)))))....(....).. ( -29.40) >DroPer_CAF1 32154 102 + 1 UGCUUUCUGGGUGGAGCCCCGGCAACACCUUUGCGGACGCUGCUCUCCUCCAGAGGUGGAUUUUC---UCCCUCGAGGAGUUGCCUCG----GAGUCUGAUGGAAAUCG .(.((((((((((..((....))..)))))...(((((.((((.((((((..((((..(.....)---..))))))))))..))...)----).)))))..))))).). ( -38.70) >consensus UGCAUUUUGGGUGGAGCCUGGGCAACACCUUUGCGGAUCCC_______________UGGAUUUUCAUCUCCCUAUUGG__AGGCAUCG____GAUUCUGAUGGAAACUA .(((....(((((..((....))..))))).))).((((((................(((........)))..........))))))...................... (-17.71 = -18.44 + 0.72)

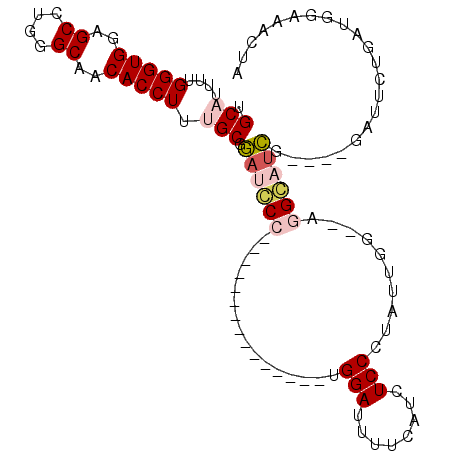
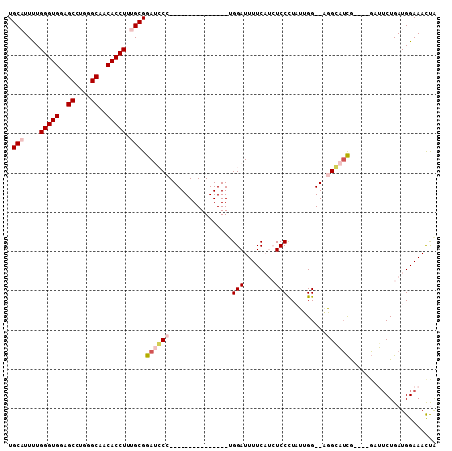
| Location | 372,263 – 372,355 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 73.96 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -13.68 |
| Energy contribution | -14.90 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 372263 92 - 23771897 UAGUUUCCAUCAGAAUAUAUAUAUUCCCU--CCAAUAAGGAGAUGAAAAUCCA---------------GGGAUUCGCAAAGGUGUUGCCCAGGCCCCACCCAAAAUGCA ..(..(((....(((((....))))).((--((.....))))...........---------------.)))..)(((..((((..((....))..)))).....))). ( -20.90) >DroPse_CAF1 31165 102 - 1 CGAUUUCCAUCAGACUC----CGAGGCAACUCCUCGAGGGA---GAAAAUCCACCUCUGGAGGAGAGCAGCGUCCGCAAAGGUGUUGCCGGGGCUCCACCCAGAAAGCA ...(((((..(((((((----.((((.....)))))))(((---.....)))...))))..)))))((((((.((.....)))))))).(((......)))........ ( -36.50) >DroSec_CAF1 26359 88 - 1 UAGUUUCCAUCAGAAGC----CGAUGCCU--CCAAUAAGGAGAUGAAAAUCCG---------------GGGAUCCGCAAAGGUGUUGCCCAGGCCCCACCCAAAAUGCA ..(((((.....)))))----.(((.(((--((.....)))(((....)))..---------------)).))).(((..((((..((....))..)))).....))). ( -21.80) >DroSim_CAF1 27169 88 - 1 UAGUUUCCAUCAGAAGC----CGAUCCCU--CCAAUAAGGAGAUGAAAAUCCA---------------GGGAUCCGCAAAGGUGUUGCCCAGGCCCCACCCAAAAUGCA ..(((((.....)))))----.(((((((--((.....)))(((....)))..---------------)))))).(((..((((..((....))..)))).....))). ( -26.70) >DroYak_CAF1 28107 88 - 1 UAGUUUCCAUCAGCAGC----CGAUCCCA--CCAAUAGGGAGAUGAAAAUCCA---------------GGGAUCCGCAAAGGUGUUGCCCAGGCUCCACCCAAAAUGCA ............(((((----.((((((.--((....))(.(((....)))).---------------)))))).))...((((..((....))..)))).....))). ( -30.30) >DroPer_CAF1 32154 102 - 1 CGAUUUCCAUCAGACUC----CGAGGCAACUCCUCGAGGGA---GAAAAUCCACCUCUGGAGGAGAGCAGCGUCCGCAAAGGUGUUGCCGGGGCUCCACCCAGAAAGCA ...(((((..(((((((----.((((.....)))))))(((---.....)))...))))..)))))((((((.((.....)))))))).(((......)))........ ( -36.50) >consensus UAGUUUCCAUCAGAAGC____CGAUCCCA__CCAAUAAGGAGAUGAAAAUCCA_______________GGGAUCCGCAAAGGUGUUGCCCAGGCCCCACCCAAAAUGCA ......................((((((..........(((........))).(((....))).....)))))).(((..((((..((....))..)))).....))). (-13.68 = -14.90 + 1.22)

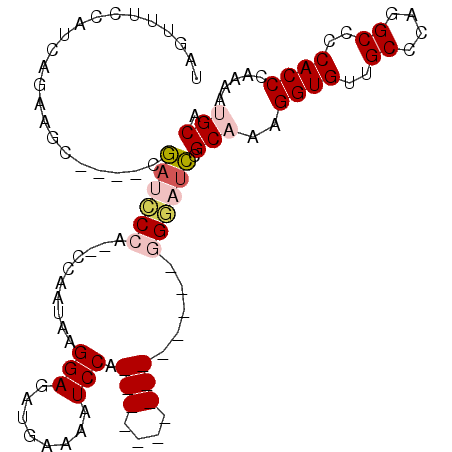
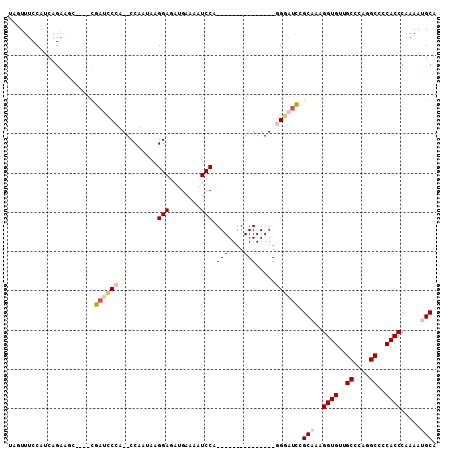
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:40 2006