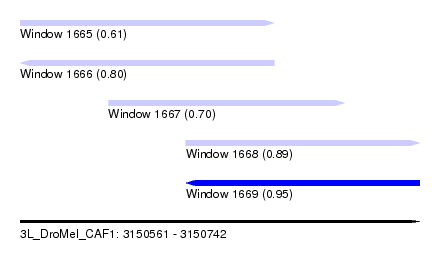
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,150,561 – 3,150,742 |
| Length | 181 |
| Max. P | 0.951607 |

| Location | 3,150,561 – 3,150,676 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 92.46 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -30.92 |
| Energy contribution | -32.03 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3150561 115 + 23771897 AAUGGUGAAAGUUGGCGCAGUUUGGAUUUGGAUGAGGGGCAGGUCAUUGUACACCAGCUCAAAAAGUUGAGCGCCUAUUGUCGUCUGAUAUUCAGACAUGUAAAGACAAUUGCUC ...((..(..(((...((((((((((((..(((((.((((.(((........))).((((((....))))))))))....)))))..).)))))))).)))...)))..)..)). ( -38.20) >DroSec_CAF1 14350 115 + 1 AAUGGUGAAAGUUGGCGCAGUUUGGAUUUGGAUGAGGGGCAGGUCAUUGUACACCAGCUCCAAAAGUUGAGCGCCUGUCGUCGUCUGAUAUUCAGACAUGUAAAGACAAUUGCUC ................((((((.(..(((((((((.((((.(((........))).((((........)))))))).)))))(((((.....)))))...))))..))))))).. ( -38.10) >DroEre_CAF1 14471 112 + 1 AAUGGUGAAAGUUGGCGCAGUUUGGAAUUGGAUGAGGGGCAGGUCAUUAUACUCCAGGUCCAAAAGUUGAGCGCCU---GUCGUCUGAUCGUCAGACAUGUAAAAACGAUUGCUC ...((..(..(((...(((((((((.((..(((((.((((.((..........))..(..((.....))..)))))---.)))))..))..)))))).)))...)))..)..)). ( -32.60) >consensus AAUGGUGAAAGUUGGCGCAGUUUGGAUUUGGAUGAGGGGCAGGUCAUUGUACACCAGCUCCAAAAGUUGAGCGCCU_U_GUCGUCUGAUAUUCAGACAUGUAAAGACAAUUGCUC ...((..(..(((...((((((((((((..(((((.((((.(((........))).((((........))))))))....)))))..).)))))))).)))...)))..)..)). (-30.92 = -32.03 + 1.11)

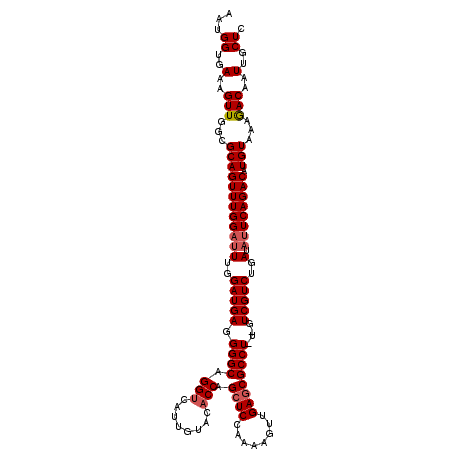
| Location | 3,150,561 – 3,150,676 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 92.46 |
| Mean single sequence MFE | -28.07 |
| Consensus MFE | -25.34 |
| Energy contribution | -26.01 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3150561 115 - 23771897 GAGCAAUUGUCUUUACAUGUCUGAAUAUCAGACGACAAUAGGCGCUCAACUUUUUGAGCUGGUGUACAAUGACCUGCCCCUCAUCCAAAUCCAAACUGCGCCAACUUUCACCAUU ..(((((((((.......(((((.....))))))))))).(((((((((....)))))).(((........))).)))..................)))................ ( -26.71) >DroSec_CAF1 14350 115 - 1 GAGCAAUUGUCUUUACAUGUCUGAAUAUCAGACGACGACAGGCGCUCAACUUUUGGAGCUGGUGUACAAUGACCUGCCCCUCAUCCAAAUCCAAACUGCGCCAACUUUCACCAUU (((....((((......((((((.....))))))..))))(((((......((((((...((.(((........))).))...))))))........)))))....)))...... ( -30.04) >DroEre_CAF1 14471 112 - 1 GAGCAAUCGUUUUUACAUGUCUGACGAUCAGACGAC---AGGCGCUCAACUUUUGGACCUGGAGUAUAAUGACCUGCCCCUCAUCCAAUUCCAAACUGCGCCAACUUUCACCAUU ((....)).........((((((.....))))))..---.(((((......((((((..((((....................))))..))))))..)))))............. ( -27.45) >consensus GAGCAAUUGUCUUUACAUGUCUGAAUAUCAGACGAC_A_AGGCGCUCAACUUUUGGAGCUGGUGUACAAUGACCUGCCCCUCAUCCAAAUCCAAACUGCGCCAACUUUCACCAUU (((..............((((((.....))))))......(((((......((((((...((.(((........))).))...))))))........)))))....)))...... (-25.34 = -26.01 + 0.67)

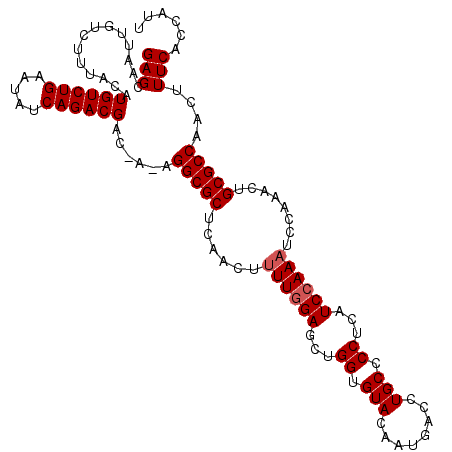
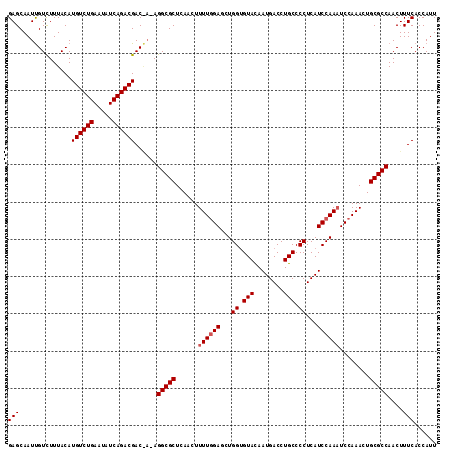
| Location | 3,150,601 – 3,150,708 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 90.65 |
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -24.85 |
| Energy contribution | -25.30 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3150601 107 + 23771897 AGGUCAUUGUACACCAGCUCAAAAAGUUGAGCGCCUAUUGUCGUCUGAUAUUCAGACAUGUAAAGACAAUUGCUCAAUUGCAUUAGCAGUUGUCGUCAUACGAAUAC .(((........))).((((((....))))))((..(((((((((((.....))))).(....))))))).)).(((((((....)))))))(((.....))).... ( -30.60) >DroSec_CAF1 14390 107 + 1 AGGUCAUUGUACACCAGCUCCAAAAGUUGAGCGCCUGUCGUCGUCUGAUAUUCAGACAUGUAAAGACAAUUGCUCAAUUGCUUUGGCAGUUGUCGUCAUACGAAUAC ......((((((..(((((((((((((((((((..((((...(((((.....))))).......))))..)))))))))..))))).)))))..))...)))).... ( -32.90) >DroEre_CAF1 14511 104 + 1 AGGUCAUUAUACUCCAGGUCCAAAAGUUGAGCGCCU---GUCGUCUGAUCGUCAGACAUGUAAAAACGAUUGCUCAAUUGCUUUGGCAAUUGUCGUCAUACGAAUAC ..............(((..((((((((((((((...---((((((((.....)))))..((....))))))))))))))..)))))...)))(((.....))).... ( -24.10) >consensus AGGUCAUUGUACACCAGCUCCAAAAGUUGAGCGCCU_U_GUCGUCUGAUAUUCAGACAUGUAAAGACAAUUGCUCAAUUGCUUUGGCAGUUGUCGUCAUACGAAUAC ..............(((((((((.((((((((..........(((((.....))))).(((....)))...))))))))...)))).)))))(((.....))).... (-24.85 = -25.30 + 0.45)

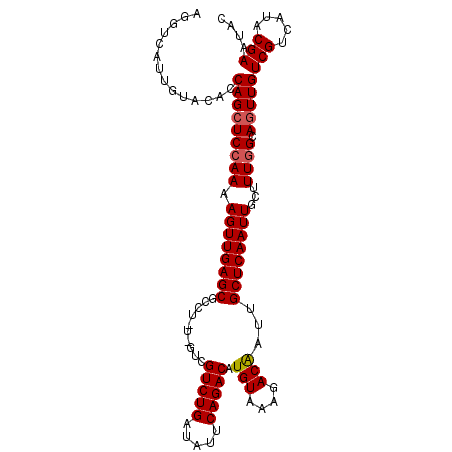
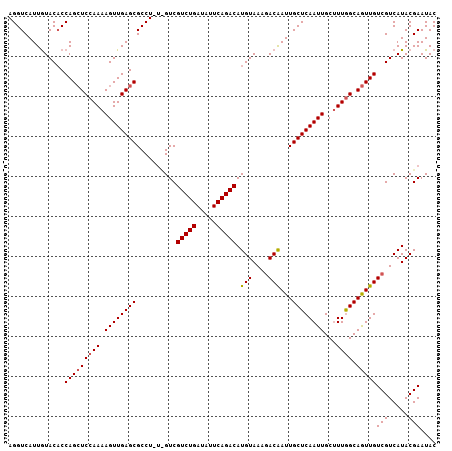
| Location | 3,150,636 – 3,150,742 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 91.19 |
| Mean single sequence MFE | -25.87 |
| Consensus MFE | -22.21 |
| Energy contribution | -23.10 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3150636 106 + 23771897 UAUUGUCGUCUGAUAUUCAGACAUGUAAAGACAAUUGCUCAAUUGCAUUAGCAGUUGUCGUCAUACGAAUACAAUUGCCAUAUCUUAUCUGAGUUUCCACCGAAAA .....(((...((.(((((((...(((..(.((((((..(((((((....)))))))(((.....)))...)))))))..)))....))))))).))...)))... ( -23.60) >DroSec_CAF1 14425 106 + 1 UGUCGUCGUCUGAUAUUCAGACAUGUAAAGACAAUUGCUCAAUUGCUUUGGCAGUUGUCGUCAUACGAAUACAAUUGCCAUAUCUUAUCUGAGUUUCUACCGAAAA .....(((...((.(((((((.....((((.(((((....)))))))))(((((((((..((....))..)))))))))........))))))).))...)))... ( -29.90) >DroEre_CAF1 14546 103 + 1 U---GUCGUCUGAUCGUCAGACAUGUAAAAACGAUUGCUCAAUUGCUUUGGCAAUUGUCGUCAUACGAAUACAAUUGCCAUAUCUUAUCCGAAUUUCCACCGAAAA .---.(((((((.....))))).........((...((......))..((((((((((..((....))..)))))))))).........))..........))... ( -24.10) >consensus U_U_GUCGUCUGAUAUUCAGACAUGUAAAGACAAUUGCUCAAUUGCUUUGGCAGUUGUCGUCAUACGAAUACAAUUGCCAUAUCUUAUCUGAGUUUCCACCGAAAA .....(((...((.(((((((..........(((((....)))))...((((((((((..((....))..)))))))))).......))))))).))...)))... (-22.21 = -23.10 + 0.89)

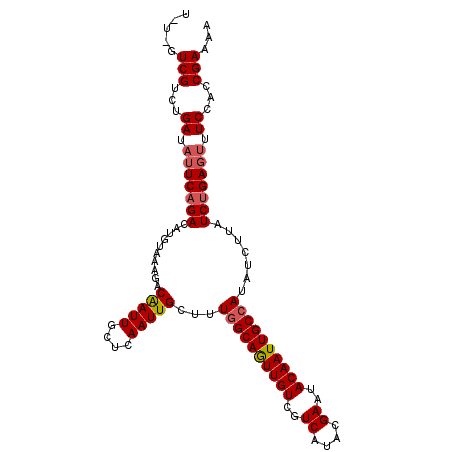
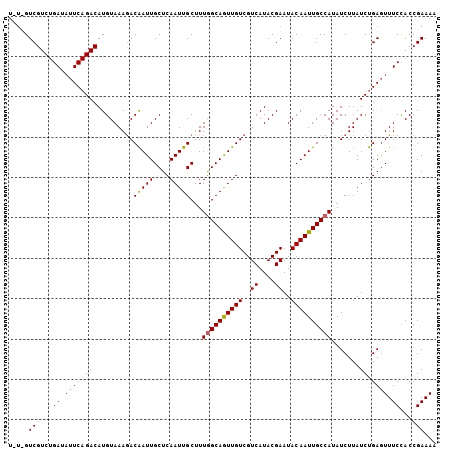
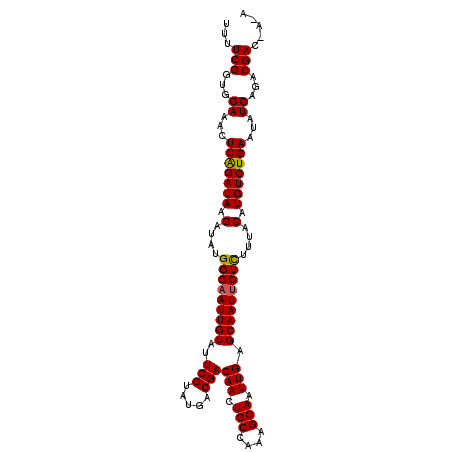
| Location | 3,150,636 – 3,150,742 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 91.19 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -25.77 |
| Energy contribution | -25.67 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3150636 106 - 23771897 UUUUCGGUGGAAACUCAGAUAAGAUAUGGCAAUUGUAUUCGUAUGACGACAACUGCUAAUGCAAUUGAGCAAUUGUCUUUACAUGUCUGAAUAUCAGACGACAAUA ...(((...((.(.(((((((.(....(((((((((..(((.....)))(((.(((....))).))).)))))))))....).))))))).).))...)))..... ( -28.50) >DroSec_CAF1 14425 106 - 1 UUUUCGGUAGAAACUCAGAUAAGAUAUGGCAAUUGUAUUCGUAUGACGACAACUGCCAAAGCAAUUGAGCAAUUGUCUUUACAUGUCUGAAUAUCAGACGACGACA ...((((((((...((......))...(((((((((..(((.....)))(((.(((....))).))).)))))))))))))).((((((.....)))))).))).. ( -26.90) >DroEre_CAF1 14546 103 - 1 UUUUCGGUGGAAAUUCGGAUAAGAUAUGGCAAUUGUAUUCGUAUGACGACAAUUGCCAAAGCAAUUGAGCAAUCGUUUUUACAUGUCUGACGAUCAGACGAC---A ......((((((((...(((......((((((((((..((....))..))))))))))..((......)).))))))))))).((((((.....))))))..---. ( -28.50) >consensus UUUUCGGUGGAAACUCAGAUAAGAUAUGGCAAUUGUAUUCGUAUGACGACAACUGCCAAAGCAAUUGAGCAAUUGUCUUUACAUGUCUGAAUAUCAGACGAC_A_A ...(((...((...(((((((.(....(((((((((..(((.....)))(((.(((....))).))).)))))))))....).)))))))...))...)))..... (-25.77 = -25.67 + -0.11)

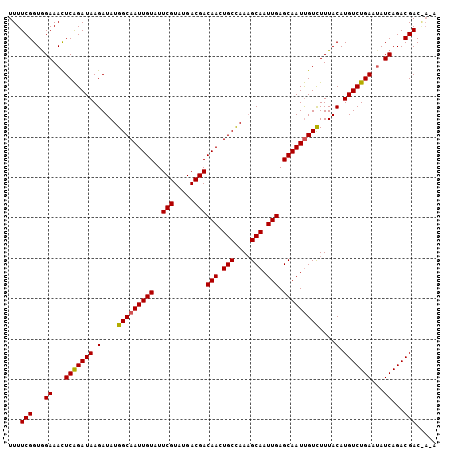
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:29 2006