| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,150,394 – 3,150,527 |
| Length | 133 |
| Max. P | 0.982819 |

| Location | 3,150,394 – 3,150,494 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 82.41 |
| Mean single sequence MFE | -31.24 |
| Consensus MFE | -25.35 |
| Energy contribution | -25.47 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3150394 100 + 23771897 AUCUGAUGUACAUAGCUGUGCU----UCCGCCAAGUGCAGCUGGUUUCAGCC----GUUGGCGUUCUGGCCAAGACCAAGACAUCUGGUUAUCUGACUUGGCCAUAUC ...(((...((.((((((..((----(.....)))..)))))))).)))(((----...)))....((((((((((((.(....))))).......)))))))).... ( -33.41) >DroSec_CAF1 14175 108 + 1 AUCUGAUGUGAAUAGCUUUGCUUAUUUCCGCCAAGUGCAGCUGGUUUCAGCCGAUCGUUGGCGUUCUGGCCAAGACCAAGACAUCUGGUUAUCUGACUUGGUCAUAUC ....(((((((..(((...)))........((((((.(((.(((((...(((((...))))).....))))).(((((.(....))))))..)))))))))))))))) ( -30.00) >DroEre_CAF1 14305 95 + 1 AUCUG---UGGA----------UAUUUCCGCCAAGUGCAGCAGGUUUUAGCCGUUCGUUGGCGUUCUGGCCAAGACCAACACAUCUGGUUAUCUGACUUGGCCAUAUC .....---.(((----------....)))(((((((.(((..((((...((((.....)))).....))))..(((((.......)))))..))))))))))...... ( -30.30) >consensus AUCUGAUGUGAAUAGCU_UGCUUAUUUCCGCCAAGUGCAGCUGGUUUCAGCCG_UCGUUGGCGUUCUGGCCAAGACCAAGACAUCUGGUUAUCUGACUUGGCCAUAUC .............................(((((((.(((..((((...((((.....)))).....))))..(((((.......)))))..))))))))))...... (-25.35 = -25.47 + 0.11)

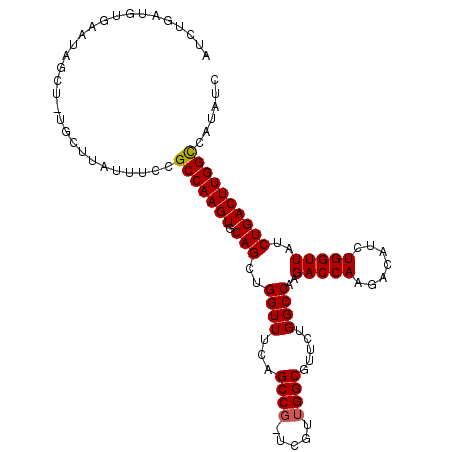
| Location | 3,150,394 – 3,150,494 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.41 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -27.53 |
| Energy contribution | -29.20 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3150394 100 - 23771897 GAUAUGGCCAAGUCAGAUAACCAGAUGUCUUGGUCUUGGCCAGAACGCCAAC----GGCUGAAACCAGCUGCACUUGGCGGA----AGCACAGCUAUGUACAUCAGAU ....(((((((((((((((......)))).))).))))))))...(((((((----(((((....))))))...))))))((----.(.(((....))).).)).... ( -37.00) >DroSec_CAF1 14175 108 - 1 GAUAUGACCAAGUCAGAUAACCAGAUGUCUUGGUCUUGGCCAGAACGCCAACGAUCGGCUGAAACCAGCUGCACUUGGCGGAAAUAAGCAAAGCUAUUCACAUCAGAU (((..(((((((.((..........)).))))))).......(((((((((....((((((....))))))...))))))......(((...))).)))..))).... ( -31.40) >DroEre_CAF1 14305 95 - 1 GAUAUGGCCAAGUCAGAUAACCAGAUGUGUUGGUCUUGGCCAGAACGCCAACGAACGGCUAAAACCUGCUGCACUUGGCGGAAAUA----------UCCA---CAGAU ((((((((((((((((...((.....)).)))).)))))))....((((((.(..((((........))))..)))))))...)))----------))..---..... ( -32.10) >consensus GAUAUGGCCAAGUCAGAUAACCAGAUGUCUUGGUCUUGGCCAGAACGCCAACGA_CGGCUGAAACCAGCUGCACUUGGCGGAAAUAAGCA_AGCUAUCCACAUCAGAU ....(((((((((((((((......)))).))).))))))))...((((((....((((((....))))))...))))))............................ (-27.53 = -29.20 + 1.67)

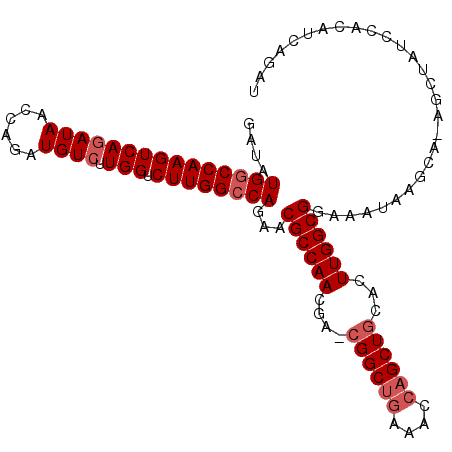
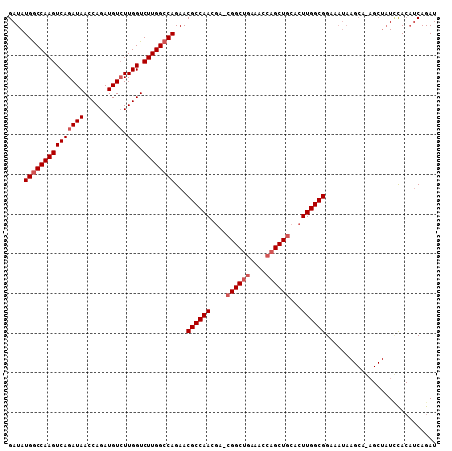
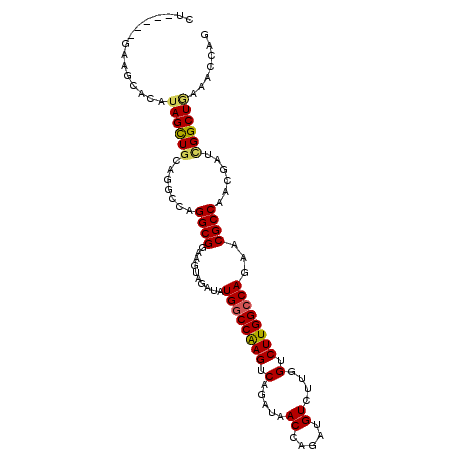
| Location | 3,150,430 – 3,150,527 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 80.52 |
| Mean single sequence MFE | -30.68 |
| Consensus MFE | -20.25 |
| Energy contribution | -20.44 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3150430 97 - 23771897 CU-----GAAGCGCAUAGCUGCAGGCCAGGCGGAAGUAGAUAUGGCCAAGUCAGAUAACCAGAUGUCUUGGUCUUGGCCAGAACGCCAAC----GGCUGAAACCAG ((-----(.(((.....))).)))(((.((((..........(((((((((((((((......)))).))).))))))))...))))...----)))......... ( -34.02) >DroSec_CAF1 14215 101 - 1 CU-----GAAGCACAUAGCUGCAGGCCAGGCGGAAGUAGAUAUGACCAAGUCAGAUAACCAGAUGUCUUGGUCUUGGCCAGAACGCCAACGAUCGGCUGAAACCAG ((-----(.(((.....))).)))(((..((....)).(((..(((((((.((..........)).)))))))(((((......)))))..))))))......... ( -32.10) >DroEre_CAF1 14332 101 - 1 CU-----GAAGCACAUAGCUGCAGGCCAGGCGGAAGUAGAUAUGGCCAAGUCAGAUAACCAGAUGUGUUGGUCUUGGCCAGAACGCCAACGAACGGCUAAAACCUG ((-----((.(((......))).(((((.((....)).....)))))...)))).....(((..((.(((((....))))).))(((.......)))......))) ( -30.60) >DroAna_CAF1 13578 89 - 1 CCACAUGGAAACUCAAAGUUGCA-----GGCGGAAGUAGAUAUGGCCGAG------AACCAGAUGU------CUUGGCCAGAACGCCAACAGUUGGCUGAAAGCUG ......(....)........((.-----((((..........((((((((------(........)------))))))))...))))..(((....)))...)).. ( -26.02) >consensus CU_____GAAGCACAUAGCUGCAGGCCAGGCGGAAGUAGAUAUGGCCAAGUCAGAUAACCAGAUGUCUUGGUCUUGGCCAGAACGCCAACGAUCGGCUGAAACCAG ...............((((((.......((((..........((((((((.(.....((.....))....).))))))))...))))......))))))....... (-20.25 = -20.44 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:24 2006