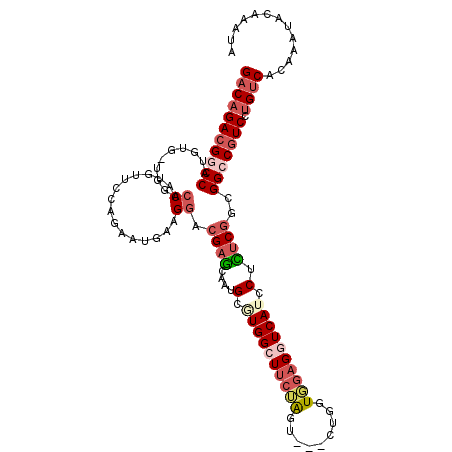
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,141,145 – 3,141,255 |
| Length | 110 |
| Max. P | 0.854058 |

| Location | 3,141,145 – 3,141,255 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.53 |
| Mean single sequence MFE | -35.38 |
| Consensus MFE | -19.08 |
| Energy contribution | -19.58 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
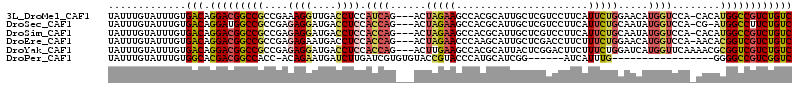
>3L_DroMel_CAF1 3141145 110 + 23771897 UAUUUGUAUUUGUGACAGGACGGCCGCCGAAAGGUUGACCUCCAUCAG---ACUAGAAGCCACGCAUUGCUCGUCCUUCAUUCUGGAACAUGGUCCA-CACAUGGCCGUCUGUC .............(((((.(((((((((....))).((((....((((---(...((((..(((.......))).))))..))))).....))))..-.....))))))))))) ( -36.60) >DroSec_CAF1 5121 109 + 1 UAUUUGUAUUUGUGACAGGAUGGCCGCCGAGAGGAUGACCUCCACCAG---ACUAGAAGCCACGCAUUGCUCGUCCUUCAUUCUGCAAUAUGGUCCA-CG-AUGGCCUUCUGUC .............(((((((.(((((((....))..((((.....(((---(...((((..(((.......))).))))..))))......))))..-..-.)))))))))))) ( -31.40) >DroSim_CAF1 5110 110 + 1 UAUUUGUAUUUGUGACAGGACGGCCGCCGAGAGGAUGACCUCCACCAG---ACUAGAAGCCACGCAUUGCUCGUCCUUCAUUCUGCAAUAUGGUCCA-CACAUGGCCGUCUGUC .............(((((.(((((((((....))..((((.....(((---(...((((..(((.......))).))))..))))......))))..-....)))))))))))) ( -33.50) >DroEre_CAF1 5125 110 + 1 UAUUUGUAUUUGUGACAGGACGGCCGCCGAGAGAAUGACCUCCACCAG---ACUAGAACCCAAGCAUUGCUCGACCUUCUUUCUGGAACAUGGUCCA-AACACGGUCGUCUGUC .............(((((.(((((((.(....)...((((....((((---(..((((..(.(((...))).)...)))).))))).....))))..-....)))))))))))) ( -32.70) >DroYak_CAF1 5148 111 + 1 UAUUUGUAUUUGUGACAGGACGGCCGCCGAGAGGAUGACCUCCACCAG---ACUUGAAGCCACGCAUUACUCGGACUUCUUUCUGGAUCAUGGUUCAAAACGCGGUCGUCUGUC .............(((((.((((((((.(.((((....))))).((((---(...((((((...........)).))))..)))))...............))))))))))))) ( -38.80) >DroPer_CAF1 5275 90 + 1 UAUUUGUAUUUGUGGCACGACGGCCACC-ACAGAAUGAUCUUGAUCGUGUGUACCGUACCCAUGCAUCGG------AUCAUUUG-----------------GGGGCCGUCGGUC .............(((.((((((((.((-...(((((((((.(((((((.(((...))).)))).)))))------))))))).-----------------))))))))))))) ( -39.30) >consensus UAUUUGUAUUUGUGACAGGACGGCCGCCGAGAGGAUGACCUCCACCAG___ACUAGAAGCCACGCAUUGCUCGUCCUUCAUUCUGGAACAUGGUCCA_CACAUGGCCGUCUGUC .............(((.(((((((((....((((....)))).((((......(((((......................))))).....))))........)))))))))))) (-19.08 = -19.58 + 0.51)
| Location | 3,141,145 – 3,141,255 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.53 |
| Mean single sequence MFE | -36.32 |
| Consensus MFE | -21.42 |
| Energy contribution | -23.21 |
| Covariance contribution | 1.79 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
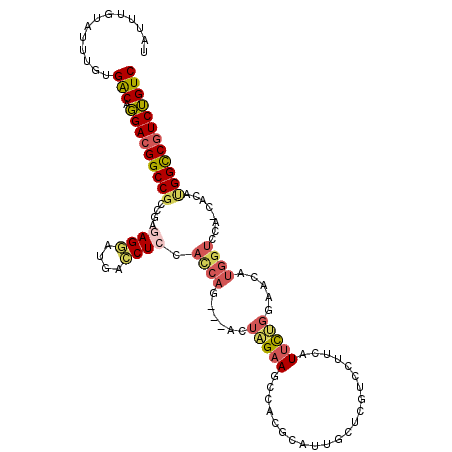
>3L_DroMel_CAF1 3141145 110 - 23771897 GACAGACGGCCAUGUG-UGGACCAUGUUCCAGAAUGAAGGACGAGCAAUGCGUGGCUUCUAGU---CUGAUGGAGGUCAACCUUUCGGCGGCCGUCCUGUCACAAAUACAAAUA (((((((((((...(.-((((......)))).).((((((((((....).)))((((((((..---....))))))))...))))))..)))))).)))))............. ( -39.80) >DroSec_CAF1 5121 109 - 1 GACAGAAGGCCAU-CG-UGGACCAUAUUGCAGAAUGAAGGACGAGCAAUGCGUGGCUUCUAGU---CUGGUGGAGGUCAUCCUCUCGGCGGCCAUCCUGUCACAAAUACAAAUA (((((..((((..-((-.(((...((((((..............))))))..(((((((((..---....))))))))))))...))..))))...)))))............. ( -34.14) >DroSim_CAF1 5110 110 - 1 GACAGACGGCCAUGUG-UGGACCAUAUUGCAGAAUGAAGGACGAGCAAUGCGUGGCUUCUAGU---CUGGUGGAGGUCAUCCUCUCGGCGGCCGUCCUGUCACAAAUACAAAUA (((((((((((.((.(-.(((...((((((..............))))))..(((((((((..---....)))))))))))).).))..)))))).)))))............. ( -38.54) >DroEre_CAF1 5125 110 - 1 GACAGACGACCGUGUU-UGGACCAUGUUCCAGAAAGAAGGUCGAGCAAUGCUUGGGUUCUAGU---CUGGUGGAGGUCAUUCUCUCGGCGGCCGUCCUGUCACAAAUACAAAUA ((((((((.((((...-..((((.....(((((.((((..((((((...)))))).))))..)---))))....)))).........)))).))).)))))............. ( -35.24) >DroYak_CAF1 5148 111 - 1 GACAGACGACCGCGUUUUGAACCAUGAUCCAGAAAGAAGUCCGAGUAAUGCGUGGCUUCAAGU---CUGGUGGAGGUCAUCCUCUCGGCGGCCGUCCUGUCACAAAUACAAAUA ((((((((.((((...............(((((..(((((((((....).)).))))))...)---)))).(((((....)))))..)))).))).)))))............. ( -40.70) >DroPer_CAF1 5275 90 - 1 GACCGACGGCCCC-----------------CAAAUGAU------CCGAUGCAUGGGUACGGUACACACGAUCAAGAUCAUUCUGU-GGUGGCCGUCGUGCCACAAAUACAAAUA ...((((((((((-----------------((((((((------(.(((.(.((.(((...))).)).))))..))))))).)).-)).))))))))................. ( -29.50) >consensus GACAGACGGCCAUGUG_UGGACCAUGUUCCAGAAUGAAGGACGAGCAAUGCGUGGCUUCUAGU___CUGGUGGAGGUCAUCCUCUCGGCGGCCGUCCUGUCACAAAUACAAAUA (((((((((((..........((...............)).((((....(.((((((((((.........)))))))))).).))))..))))))).))))............. (-21.42 = -23.21 + 1.79)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:17 2006