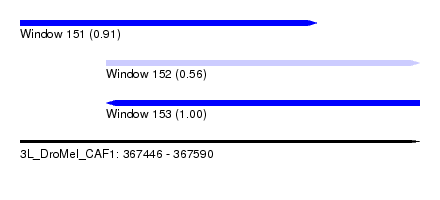
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 367,446 – 367,590 |
| Length | 144 |
| Max. P | 0.998084 |

| Location | 367,446 – 367,553 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.23 |
| Mean single sequence MFE | -23.83 |
| Consensus MFE | -7.51 |
| Energy contribution | -9.07 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.32 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 367446 107 + 23771897 ---AUAUUAUUUACCCUG------AACUACCUUCAAUGGCUA---GUUUUAUUGGGUCUGCCAGAUUGUUUU-UAAGGAUUGUUAAAUACAAACAUCCUAAUACUAGCAAUCCCAAUGAA ---...............------(((((((......)).))---)))((((((((..(((.((..((((..-..(((((.(((.......)))))))))))))).)))..)))))))). ( -24.10) >DroSec_CAF1 21558 105 + 1 ---AUAAUAUUUACCCUG------AACUACCAUCAAUGGCUG---AU-UAAUUG-GUCUGCCAGAUUGAUUU-UAGGGAUUGUUAAAUACAGAGAUCCUAAUACUAGCAAUCACAUUGAA ---...........((((------((.....(((((((((.(---((-(....)-))).))))..)))))))-))))((((((((.....((.....)).....))))))))........ ( -22.20) >DroSim_CAF1 22373 106 + 1 ---AUAAUAUUUACCCUG------AACUACCUUCAAUGGCUG---GU-CAAUUGGGUCUGCCAGAUUGAUUU-UAGGGAUUGUUAAACACAGACAUCCUAAUACUAGCAAUCACAUUGAA ---...............------........((((((((((---((-(((((.((....)).))))))...-..(((((.(((.......))))))))....))))).....)))))). ( -22.90) >DroEre_CAF1 23029 104 + 1 GGUGUAUUA-UUAGCCUG------AAAUACCUUCAAACCCAG---GC-UUAUUGAGCCUGGUAGAUUGUUUC-UAGGGAUUGUUAAAUAC----AUUUUAAUACUAGCAAUCACAUCGAA (((((....-....((((------(((((...((....((((---((-((...))))))))..)).))))))-.)))((((((((.....----..........)))))))))))))... ( -30.16) >DroYak_CAF1 23213 114 + 1 UGUUUAUUA-UUACCCUUAUUAUUAACUGCCUUCAAACGCAGAUUGU-UUAUUUGGUCUGCCAGAUUGUUUUUUCGGUAUUGUUAAAUAC----ACUUUAAUACUAGCAAUCACAUUGAG .........-......................((((..((((((((.-.....))))))))..(((((((.....(((((((........----....))))))))))))))...)))). ( -19.80) >consensus ___AUAUUAUUUACCCUG______AACUACCUUCAAUGGCUG___GU_UUAUUGGGUCUGCCAGAUUGUUUU_UAGGGAUUGUUAAAUACA_A_AUCCUAAUACUAGCAAUCACAUUGAA ................................((((((((...................))))(((((((.....((((((((.....)))...)))))......)))))))...)))). ( -7.51 = -9.07 + 1.56)

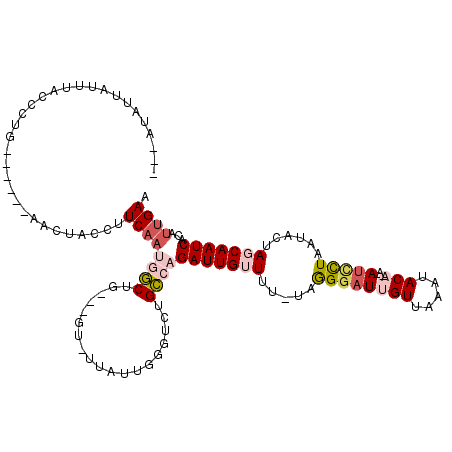
| Location | 367,477 – 367,590 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.66 |
| Mean single sequence MFE | -25.65 |
| Consensus MFE | -16.64 |
| Energy contribution | -16.60 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 367477 113 + 23771897 UA---GUUUUAUUGGGUCUGCCAGAUUGUUUU-UAAGGAUUGUUAAAUACAAACAUCCUAAUACUAGCAAUCCCAAUGAAU---CUCUAGCAUUAGCGAGUAUCUAUCAAAAGGCAGAAA ..---...........((((((.(((((((..-(((((((.(((.......))))))))..))..))))))).....((..---(((..((....)))))..))........)))))).. ( -25.00) >DroSec_CAF1 21589 114 + 1 UG---AU-UAAUUG-GUCUGCCAGAUUGAUUU-UAGGGAUUGUUAAAUACAGAGAUCCUAAUACUAGCAAUCACAUUGAAUCAUCUCUAGCAUUAGCGAGUAUCUAACAAAAGGCAGAAC ..---..-......-.(((((((((.((((((-...(((((((((.....((.....)).....)))))))).)...))))))..))).((....))...............)))))).. ( -25.50) >DroSim_CAF1 22404 112 + 1 UG---GU-CAAUUGGGUCUGCCAGAUUGAUUU-UAGGGAUUGUUAAACACAGACAUCCUAAUACUAGCAAUCACAUUGAAU---CUCUAGCAUUAGCGAGUAUCUAACAAAAGGCAGAAC ..---..-........((((((.(((((...(-(((((..((((.......))))))))))......))))).....((..---(((..((....)))))..))........)))))).. ( -24.70) >DroEre_CAF1 23062 108 + 1 AG---GC-UUAUUGAGCCUGGUAGAUUGUUUC-UAGGGAUUGUUAAAUAC----AUUUUAAUACUAGCAAUCACAUCGAAG---CCUUGGCAUUAGCGGGGAUCUAACCAGAAACAGAAA ((---((-((...))))))......(((((((-(.((((((((((.....----..........)))))))).........---.(((.((....)).)))......))))))))))... ( -27.26) >DroYak_CAF1 23252 111 + 1 AGAUUGU-UUAUUUGGUCUGCCAGAUUGUUUUUUCGGUAUUGUUAAAUAC----ACUUUAAUACUAGCAAUCACAUUGAGA---CUUUAGCAUUAGUGGGUUUCUA-UCAGAGGCAGAAA .......-........((((((.(((((((.....(((((((........----....)))))))))))))).....((((---((...((....)).))))))..-.....)))))).. ( -25.80) >consensus UG___GU_UUAUUGGGUCUGCCAGAUUGUUUU_UAGGGAUUGUUAAAUACA_A_AUCCUAAUACUAGCAAUCACAUUGAAU___CUCUAGCAUUAGCGAGUAUCUAACAAAAGGCAGAAA ................((((((.(((((((.....((((((((.....)))...)))))......)))))))............(((..((....)))))............)))))).. (-16.64 = -16.60 + -0.04)

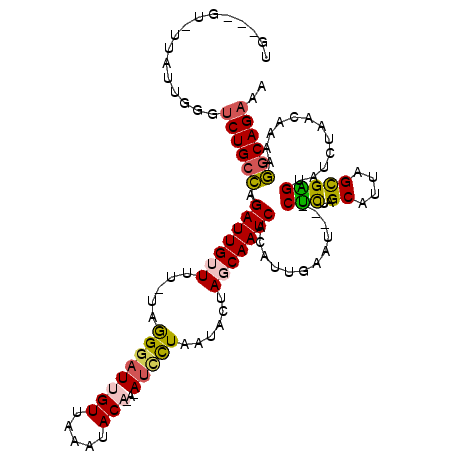
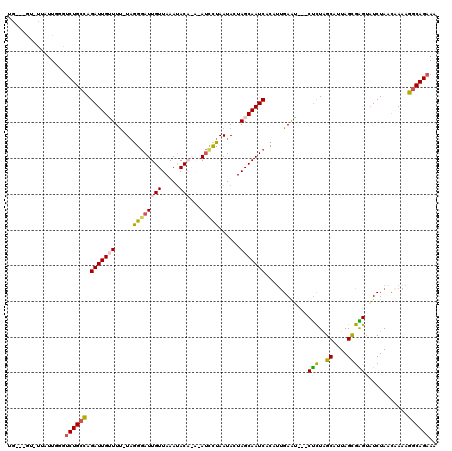
| Location | 367,477 – 367,590 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.66 |
| Mean single sequence MFE | -28.51 |
| Consensus MFE | -15.99 |
| Energy contribution | -17.99 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.56 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
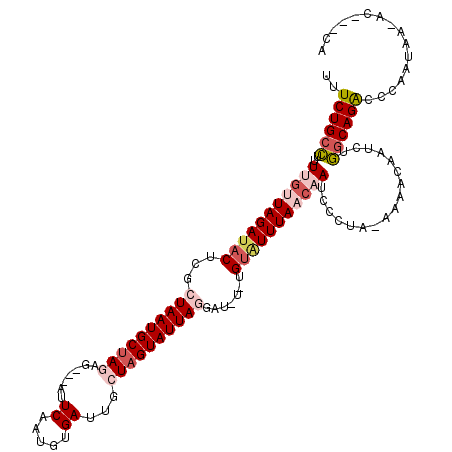
>3L_DroMel_CAF1 367477 113 - 23771897 UUUCUGCCUUUUGAUAGAUACUCGCUAAUGCUAGAG---AUUCAUUGGGAUUGCUAGUAUUAGGAUGUUUGUAUUUAACAAUCCUUA-AAAACAAUCUGGCAGACCCAAUAAAAC---UA ..((((((..((((((((((.((.((((((((((..---((((....))))..))))))))))))))))))).(((((......)))-))..)))...))))))...........---.. ( -32.20) >DroSec_CAF1 21589 114 - 1 GUUCUGCCUUUUGUUAGAUACUCGCUAAUGCUAGAGAUGAUUCAAUGUGAUUGCUAGUAUUAGGAUCUCUGUAUUUAACAAUCCCUA-AAAUCAAUCUGGCAGAC-CAAUUA-AU---CA (.((((((..(((((((((((((.((((((((((..((.((.....)).))..)))))))))))).....)))))))))))......-..........)))))).-).....-..---.. ( -32.57) >DroSim_CAF1 22404 112 - 1 GUUCUGCCUUUUGUUAGAUACUCGCUAAUGCUAGAG---AUUCAAUGUGAUUGCUAGUAUUAGGAUGUCUGUGUUUAACAAUCCCUA-AAAUCAAUCUGGCAGACCCAAUUG-AC---CA ..((((((..(((((((((((((.((((((((((..---(((......)))..)))))))))))).....)))))))))))......-..........))))))........-..---.. ( -31.07) >DroEre_CAF1 23062 108 - 1 UUUCUGUUUCUGGUUAGAUCCCCGCUAAUGCCAAGG---CUUCGAUGUGAUUGCUAGUAUUAAAAU----GUAUUUAACAAUCCCUA-GAAACAAUCUACCAGGCUCAAUAA-GC---CU ....(((((((((...((..((.((....))...))---..)).....(((((..(((((......----)))))...))))).)))-)))))).......(((((.....)-))---)) ( -24.70) >DroYak_CAF1 23252 111 - 1 UUUCUGCCUCUGA-UAGAAACCCACUAAUGCUAAAG---UCUCAAUGUGAUUGCUAGUAUUAAAGU----GUAUUUAACAAUACCGAAAAAACAAUCUGGCAGACCAAAUAA-ACAAUCU ..((((((...((-(.(........((((((((.((---((.......))))..))))))))..(.----(((((....)))))).......).))).))))))........-....... ( -22.00) >consensus UUUCUGCCUUUUGUUAGAUACUCGCUAAUGCUAGAG___AUUCAAUGUGAUUGCUAGUAUUAGGAU_U_UGUAUUUAACAAUCCCUA_AAAACAAUCUGGCAGACCCAAUAA_AC___CA ..((((((..(((((((((((...((((((((((.......((.....))...)))))))))).......))))))))))).................))))))................ (-15.99 = -17.99 + 2.00)

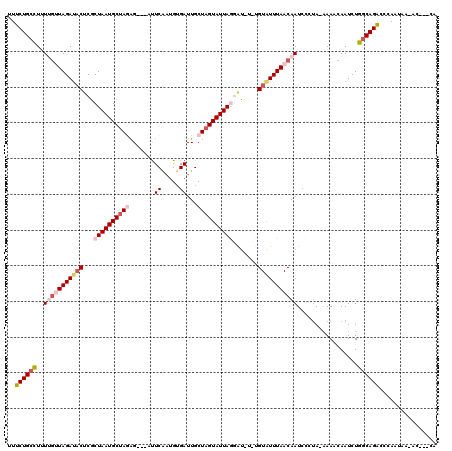
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:38 2006