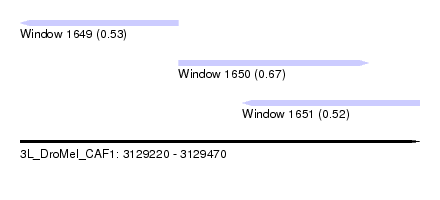
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,129,220 – 3,129,470 |
| Length | 250 |
| Max. P | 0.668094 |

| Location | 3,129,220 – 3,129,319 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 82.20 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -22.62 |
| Energy contribution | -23.48 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3129220 99 - 23771897 UCUCGCACGCCGAUUCAAUUGGUGUUGAGGAUGCUGCCCCCGCUGCGAAUUCAUCAGCUGCAGCGGCCUCCUCGACGCCAA---AUAAGACCAACUACCGCG ...(((............(((((((((((((.(((((..(.((((.(....)..)))).)..))))).)))))))))))))---...((.....))...))) ( -37.00) >DroSec_CAF1 18163 99 - 1 UCUCGCACGCCGAUUCAAUUAGUGUCGAGGAAGCUGCCCCCGCUUCAAAUUCAUCAGCUGCAGCGGCGUCCUCGGCGCCCA---AUAAGACCGACUACCGCG ...(((...............((((((((((.(((((..(.(((...........))).)..))))).))))))))))...---...((.....))...))) ( -29.70) >DroSim_CAF1 16461 99 - 1 UCUCGCACGCCGAUUCAAUUAGUGUCGAGGAAGCUGUCCCCGCUGAAAAUUCAUCAGCUGCAGCGGCGUCCUCGGCGCCCA---AUAAGACCGACUACCGCG ...(((...............((((((((((.(((((..(.(((((.......))))).)..))))).))))))))))...---...((.....))...))) ( -33.80) >DroEre_CAF1 16848 99 - 1 UCUCGCAUGCCGAUUCUAUUGGUGUUGAGGAAGUUGCCCCCGCUUCGAAUUCAUCAGCUGCAGCGGCUUCCUCGGCGCCCA---AUAAGACCGACUACCGCG ...(((..(.((.(..(((((((((((((((((((((..(.(((..((.....))))).)..))))))))))))))).)))---)))..).)).)....))) ( -36.80) >DroYak_CAF1 19488 99 - 1 UCUCACACGCCGAUUCCAUUGGUGUUGAGGAAGUUGCCCCCGCAUCGAAUUCAUCAGCUGCAGCGGCUUCCUCGGCGCCCA---AUAAGACCGACUACCGCG .......(((((.((..((((((((((((((((((((....(((.(..........).))).))))))))))))))).)))---))..)).))......))) ( -31.90) >DroAna_CAF1 16003 93 - 1 UCUCGCACGCCGAUUCCAUUGGCGUGGAAGAAGUCGCACCCGCCC---------CCGCUGCGCCAGCCUCGUCAGCCGGAAAUAGUAAGACCAACUAUAGAG (((..(((((((((...)))))))))..))).(((((....)).(---------(.((((((.......)).)))).)).........)))........... ( -26.30) >consensus UCUCGCACGCCGAUUCAAUUGGUGUUGAGGAAGCUGCCCCCGCUUCGAAUUCAUCAGCUGCAGCGGCCUCCUCGGCGCCCA___AUAAGACCGACUACCGCG ...(((..((((((...))))))((((((((.(((((..(.(((...........))).)..))))).)))))))).......................))) (-22.62 = -23.48 + 0.86)

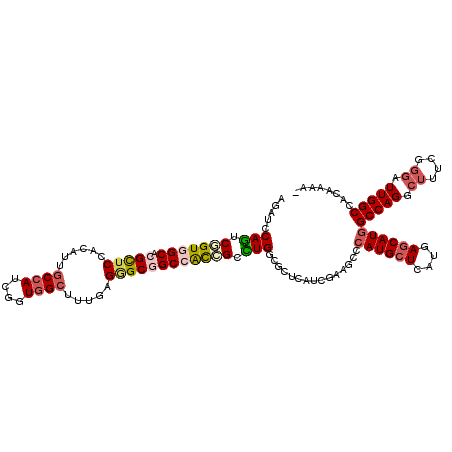
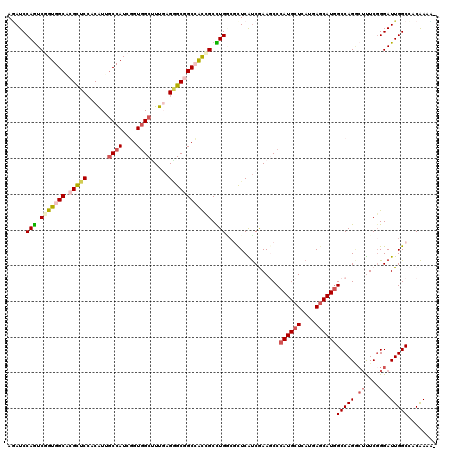
| Location | 3,129,319 – 3,129,438 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.41 |
| Mean single sequence MFE | -47.47 |
| Consensus MFE | -32.49 |
| Energy contribution | -33.43 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.668094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3129319 119 + 23771897 AGAUCCAGUCGGUGGCGCGCUCCACAUUGCCAUCGGUCGCUUUCAGAGCAGCCACCGCCUGUCUUUCGUCGAAGCCCAUGCUCAUGAGCAUGGCCAGGCUCUCGGGAUUGGCCACAAAC- ....((((((((((((..((((......((........)).....)))).))))))(((((.((((....)))).(((((((....))))))).)))))......))))))........- ( -45.70) >DroVir_CAF1 35506 120 + 1 AUAUCCAGUCGGUGGCACGUUCCACGUUGCCAUCUGUGGCUUUUAGAGCGGCGACCGCCUGGCGCUCGUCAAAGCCCAUGCUCAUGAGCAUGGCCAGGCUUUCGGGAUUGGCAGCAAAAU ....(((((((((.((.(((((......((((....)))).....))))))).)))((((((((((......))).((((((....)))))))))))))......))))))......... ( -53.70) >DroPse_CAF1 14452 119 + 1 AGAUCCAGUCUGUGGCACGCUCCACAUUCCCAUCGGUGGCCUUGAGGGCGGCCACGGCCUGGCGUUCAUCGAAUCCCAUGCUCAUGAGCAUAGCCAGGCUUUCCGGAUUGGCCACAAAC- ....((((((((((((.(((.((.((...(((....)))...)).))))))))))((((((((((((...))))...(((((....))))).))))))))....)))))))........- ( -49.80) >DroGri_CAF1 18844 119 + 1 AGAUCCAAUCUGUGGCACGUUCCACGUUGCCAUCGGUGGCUUUGAGUGCGGCCACCGCUUGCAUCUCAUCGAAGCCCAUGCUCAUCAGCAUGGCCAGACUUUCGGGAUUGGCCACAAAU- ....(((((((((((((((.....)).)))))).((((((((((((((((((....)).)))))....))))))))((((((....))))))))).........)))))))........- ( -44.60) >DroWil_CAF1 25025 119 + 1 AAAUCCAGUCGGUGGCUCGUUCAACGUUGCCAUCAGUGGCUUUGAGGGCAGCAACAGCCUGACGCUCAUCGAAUCCCAUGCUCAUCAACAUGGCCAGACUUUCGGGAUUGGCCACAAAU- ..........((((((.((.....))..)))))).((((((.((((.(((((....).))).).))))...((((((..((.(((....)))))..(.....)))))))))))))....- ( -38.10) >DroMoj_CAF1 18673 119 + 1 AGAUCCAGUCGGUGGCUCGCUCCACAUUGCCAUCGGUGGCCUUGAGGGCGGCCGUCGCUUGGCGCUCGCCAAAGCCCAUGCUCAUGAGCAUGGCCAGUGACUCGGGAUUGGCAACGAAA- ....((((((...(((.(((.((.((..((((....))))..)).))))))))((((((((((....))))).(.(((((((....))))))).).)))))....))))))........- ( -52.90) >consensus AGAUCCAGUCGGUGGCACGCUCCACAUUGCCAUCGGUGGCUUUGAGGGCGGCCACCGCCUGGCGCUCAUCGAAGCCCAUGCUCAUGAGCAUGGCCAGGCUUUCGGGAUUGGCCACAAAA_ .....(((.(((((((.(((((......((((....)))).....)))))))))))).)))...............((((((....))))))(((((.((....)).)))))........ (-32.49 = -33.43 + 0.95)
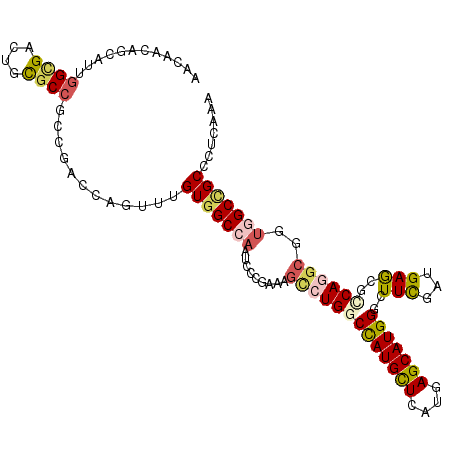
| Location | 3,129,359 – 3,129,470 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.96 |
| Mean single sequence MFE | -41.70 |
| Consensus MFE | -23.26 |
| Energy contribution | -23.73 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.56 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
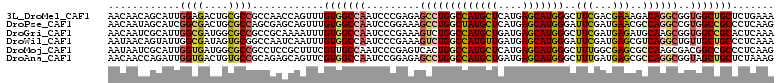
>3L_DroMel_CAF1 3129359 111 - 23771897 AACAACAGCAUUGGAGACUGCGCCGCCAACCAGUUUGUGGCCAAUCCCGAGAGCCUGGCCAUGCUCAUGAGCAUGGGCUUCGACGAAAGACAGGCGGUGGCUGCUCUGAAA .....((((.((((.((.((.(((((.((....)).))))))).))))))..(((((.(((((((....))))))).......(....).)))))....))))........ ( -42.70) >DroPse_CAF1 14492 111 - 1 AACAAUAGCAUCGGCGACUGCGCCAGCGAGCAGUUUGUGGCCAAUCCGGAAAGCCUGGCUAUGCUCAUGAGCAUGGGAUUCGAUGAACGCCAGGCCGUGGCCGCCCUCAAG ............(((((((((........))))))...(((((...((....(((((((((((((....))))))...(((...))).)))))))))))))))))...... ( -39.80) >DroGri_CAF1 18884 111 - 1 AACAAUCGCAUUGGCGAUGGCGCCGCCGCAAAAUUUGUGGCCAAUCCCGAAAGUCUGGCCAUGCUGAUGAGCAUGGGCUUCGAUGAGAUGCAAGCGGUGGCCGCACUCAAA ....(((((....))))).((((((((((.......(((((((....(....)..)))))))(((....)))....((.((.....)).))..))))))).)))....... ( -43.10) >DroWil_CAF1 25065 111 - 1 AAUAACAGUAUUGGCGAUAGUGCGGCCAAUCAAUUUGUGGCCAAUCCCGAAAGUCUGGCCAUGUUGAUGAGCAUGGGAUUCGAUGAGCGUCAGGCUGUUGCUGCCCUCAAA ............((((.....((((((.........(((((((....(....)..)))))))..(((((..(((.(....).)))..)))))))))))...))))...... ( -35.70) >DroMoj_CAF1 18713 111 - 1 AAUAAUCGCAUUGGUGAUGGCGCCGCCUCCGCUUUCGUUGCCAAUCCCGAGUCACUGGCCAUGCUCAUGAGCAUGGGCUUUGGCGAGCGCCAAGCGACGGCCGCCCUCAAG ....(((((....)))))((((..(((..(((((.(((((((((..(((......)))(((((((....)))))))...))))).))))..)))))..)))))))...... ( -43.10) >DroAna_CAF1 16136 111 - 1 AACAACCAGAUUGGUGACUGUGCCGCAGAGCAGUUCGUGGCCAAUCCGGAGAGCCUGGCCAUGCUGAUGAGCAUGGGCUUUGAUGAGCGCCAGGCGGUAGCUGCUCUAAAG .....((.(((((((((((((........))))))....))))))).))((((((((((((((((....)))))).((((....)))))))))((....)).))))).... ( -45.80) >consensus AACAACAGCAUUGGCGACUGCGCCGCCGACCAGUUUGUGGCCAAUCCCGAAAGCCUGGCCAUGCUCAUGAGCAUGGGCUUCGAUGAGCGCCAGGCGGUGGCCGCCCUCAAA ............((((....))))............(((((((.........(((((((((((((....)))))))..(((...)))..))))))..)))))))....... (-23.26 = -23.73 + 0.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:12 2006