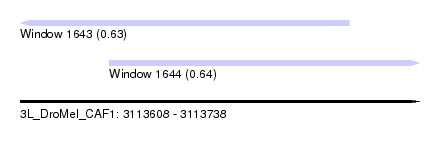
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,113,608 – 3,113,738 |
| Length | 130 |
| Max. P | 0.639156 |

| Location | 3,113,608 – 3,113,715 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 76.89 |
| Mean single sequence MFE | -35.32 |
| Consensus MFE | -21.37 |
| Energy contribution | -20.23 |
| Covariance contribution | -1.13 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
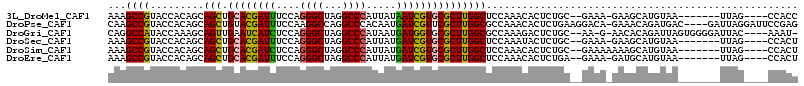
>3L_DroMel_CAF1 3113608 107 - 23771897 CGCAGAGUGUUUGGAGCCAAGCGCACGAUUAUAAUGGGCCUAGCCCUGGAAAUCGUGCAGCUGCUGUGGUACGGCUUUGGCAGUCAAAAGUGGUGAGU-AAUA---A-AUAU (((((((((((((....)))))((((((((.(...((((...))))..).)))))))).))).))))).(((.(((((........))))).)))...-....---.-.... ( -38.40) >DroVir_CAF1 2676 109 - 1 UGCAGAGUCUUUGGCGCCAAGCGCACUAUCAUUGUGGGCCUAGCCCUCGAGUUGGUUCAACUGCUUUGGUACGGCCUGGGCAGUCAGAAGUGGUAAGU-CGAAGCCU-GAG- ..(((.(.(((((((((((..((.((((...(((.((((...)))).)))..)))).).((((((..((.....))..))))))..)...))))..))-))))))))-)..- ( -38.50) >DroGri_CAF1 514 112 - 1 UGCAGAGUCUUUGGCGCCAAGCGCACCAUCAUUAUGGGCCUAGCCCUGGAGAUGAUUCAACUGCUUUGGUAUGGCCUGGGCAGUGAAAAGUGGUAAGCACAAAGCCAUUAUA ......(.(((((((((...))))(((((((((..((((...))))....)))))((((.(((((..((.....))..)))))))))...)))).....))))).)...... ( -36.10) >DroEre_CAF1 756 103 - 1 CUCAGAGUGUUUGGAGCCAAGCGCACGAUCAUAAUGGGCCUAGCCCUGGAAAUCGUGCAGCUGCUGUGGUACGGCUUUGGCAGUCAAAAGUGGUAAGU-AAUA--------U ....((.((.(..(((((.((((((((((......((((...)))).....))))))).)))((....))..)))))..))).)).............-....--------. ( -34.40) >DroWil_CAF1 745 107 - 1 UGCAGAGUAUUUGGAGCCAAACGUACAAUUAUUGUGGGCCUGGCUCUGGAAAUUGUUCAGCUGUUGUGGUAUGGCUUCGGUAGUCAAAAGUGGUAAGU-GCCA---U-UUAC ((.(.(((.((..((((((..(.((((.....)))).)..))))))..)).))).).))((((..(.((.....)).)..))))...(((((((....-))))---)-)).. ( -30.30) >DroAna_CAF1 730 105 - 1 CGCAGGACAUUUGGAGCCAAGCGCACGAUCAUUAUGGGCCUAGCCCUCGAAAUGAUCCAGAUGCUGUGGUAUGGCUUUGGUAGCCAAAAAUGGUAAGU-CGCA---G-AU-- .((..(((..(..((((((.((.((((((((((..((((...))))....)))))))........))))).))))))..)..((((....))))..))-))).---.-..-- ( -34.20) >consensus CGCAGAGUAUUUGGAGCCAAGCGCACGAUCAUUAUGGGCCUAGCCCUGGAAAUGGUUCAGCUGCUGUGGUACGGCUUUGGCAGUCAAAAGUGGUAAGU_CAAA___A_AU__ ...............((((...(.((((((.....((((...))))....)))))).).(((((((.((.....)).)))))))......)))).................. (-21.37 = -20.23 + -1.13)

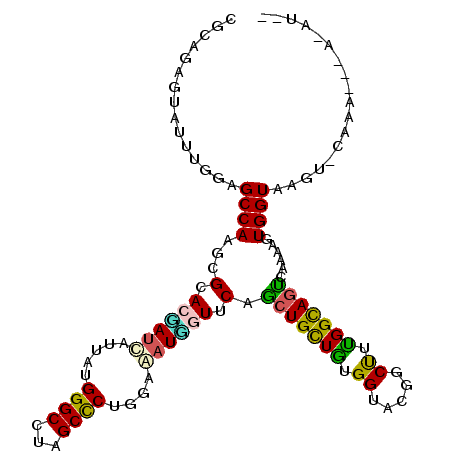
| Location | 3,113,637 – 3,113,738 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.71 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -22.25 |
| Energy contribution | -21.70 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3113637 101 + 23771897 AAAGCCGUACCACAGCAGCUGCACGAUUUCCAGGGCUAGGCCCAUUAUAAUCGUGCGCUUGGCUCCAAACACUCUGC--GAAA-GAAGCAUGUAA-------UUAG----CCACC ...((.(.....).))(((.((((((((....((((...)))).....)))))))))))(((((....(((..((.(--....-).))..)))..-------..))----))).. ( -31.70) >DroPse_CAF1 799 110 + 1 CAAGCCGUACCACAGCAGCUGUACGAUUUCCAAGGCCAGGCCCACAAUGAUCGUUCGCUUGGCGCCAAACACUCUGAAGGACA-GAAACAGAUGAC----GAUUAGGAUUCCGAG ..(((.((......)).)))...((...(((...(((((((..((.......))..))))))).........((((.....))-))..........----.....)))...)).. ( -22.90) >DroGri_CAF1 548 106 + 1 CAGGCCAUACCAAAGCAGUUGAAUCAUCUCCAGGGCUAGGCCCAUAAUGAUGGUGCGCUUGGCGCCAAAGACUCUGC--AA-G-AACACAGAUUAGUGGGGAUUAC----AAAU- ..((.....))..............((((((((((((((((((((....))))...))))))).))......((((.--..-.-....))))....)))))))...----....- ( -26.90) >DroSec_CAF1 2625 101 + 1 AAAGCCGUACCACAGCAGCUGCACGAUUUCCAGGGCUAGGCCCAUUAUGAUCGUGCGCUUGGCUCCAAAUACUCUGC--GAAA-GAAGCAUGUAA-------UUAG----CCACU ...((.(.....).))(((.((((((((....((((...)))).....)))))))))))(((((..((.(((..(((--....-...))).))).-------))))----))).. ( -30.80) >DroSim_CAF1 777 102 + 1 AAAGCCGUACCACAGCAGCUGCACGAUCUCCAGGGCUAGGCCCAUUAUGAUCGUGCGCUUGGCUCCAAACACUCUGC--GAAAAAAAGCAUGUAA-------UUAG----CCACU ...((.(.....).))(((.((((((((....((((...)))).....)))))))))))(((((....(((...(((--........))))))..-------..))----))).. ( -32.90) >DroEre_CAF1 781 101 + 1 AAAGCCGUACCACAGCAGCUGCACGAUUUCCAGGGCUAGGCCCAUUAUGAUCGUGCGCUUGGCUCCAAACACUCUGA--GAAA-GAUGCAUGUAA-------UUAG----CCACU ...((.(.....).))(((.((((((((....((((...)))).....)))))))))))(((((....(((.(((..--...)-))....)))..-------..))----))).. ( -28.80) >consensus AAAGCCGUACCACAGCAGCUGCACGAUUUCCAGGGCUAGGCCCAUUAUGAUCGUGCGCUUGGCUCCAAACACUCUGC__GAAA_GAAGCAUGUAA_______UUAG____CCACU ...((((.........(((.((((((((....((((...)))).....))))))))))))))).................................................... (-22.25 = -21.70 + -0.55)
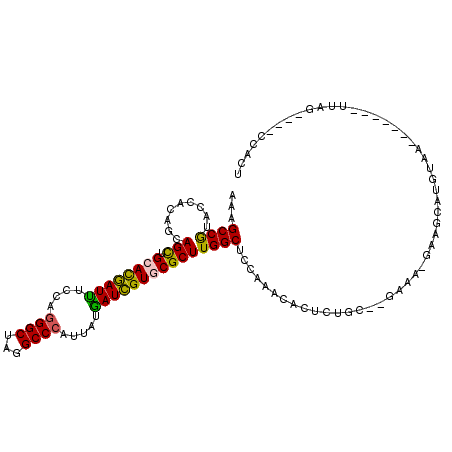
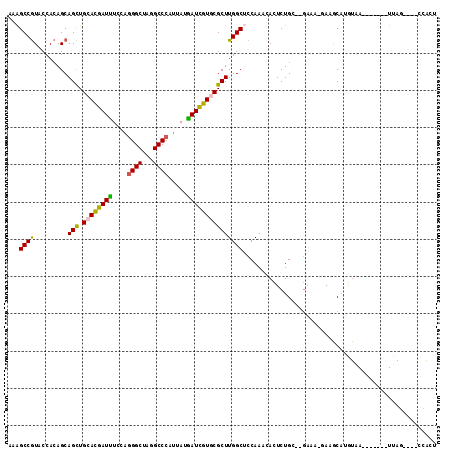
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:05 2006