| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,110,111 – 3,110,203 |
| Length | 92 |
| Max. P | 0.945747 |

| Location | 3,110,111 – 3,110,203 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 76.03 |
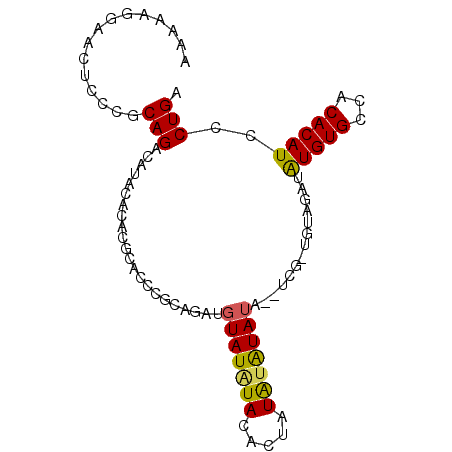
| Mean single sequence MFE | -17.30 |
| Consensus MFE | -5.34 |
| Energy contribution | -5.06 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.31 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3110111 92 - 23771897 AAAAAGGAACUCCCGCAGACAUAGACACGCACCCGCAGAUGUAUAUACACUAUACAUAUAUUUCGGUGUAGAUAUGUGCCACACAUCCCUGA .....(((......((..(((((.((((((....))..(((((((.....)))))))........))))...)))))))......))).... ( -17.50) >DroSec_CAF1 42991 84 - 1 AAAAAGGAACUCCCGCAG------ACACGCACCCGCAGAUGUAUAUACACUAUAUAUAUA--UCGCUGUAGAUAUGUGCCACACAUCCCUGA ....(((.....(..(((------.(..((....)).((((((((((.....))))))))--))))))..)..(((((...))))).))).. ( -18.50) >DroSim_CAF1 47418 90 - 1 AAAAAGGAACUCCCGCAGACAUAGACACGCACCCGCAGAUGUAUAUACACUAUAUAUAUA--UCGGUGUAGAUAUGUGCCACACAUCCCUGA .....(((......((..(((((.((((((....)).((((((((((.....))))))))--)).))))...)))))))......))).... ( -21.50) >DroEre_CAF1 47378 82 - 1 AAAAAAGAACUCCCACAGAUAUACAAACGCACUAAGA---GUAUGUAUA-CAUAUGUAUA------UGUAUAUGUGUGCCACACAUCCCUGA .............((((.(((((((.(((.((.....---)).)))(((-(....)))).------))))))).)))).............. ( -14.50) >DroYak_CAF1 47064 70 - 1 AAAAAGGAACUCCCGCAGAUGUACA----------------UAUAUACACCAUGUGUACA------UAAAUAUAUGUGCCACACAUCCCUGA .....((.....)).((((((((((----------------(((.......)))))))))------)......(((((...)))))..))). ( -14.50) >consensus AAAAAGGAACUCCCGCAGACAUACACACGCACCCGCAGAUGUAUAUACACUAUAUAUAUA__UCG_UGUAGAUAUGUGCCACACAUCCCUGA ...............(((......................(((((((.....)))))))..............(((((...)))))..))). ( -5.34 = -5.06 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:03 2006