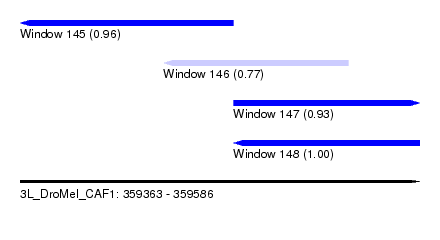
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 359,363 – 359,586 |
| Length | 223 |
| Max. P | 0.999938 |

| Location | 359,363 – 359,482 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.73 |
| Mean single sequence MFE | -38.34 |
| Consensus MFE | -34.48 |
| Energy contribution | -34.52 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 359363 119 - 23771897 -GACAAGCAAACGUCGGCUGUGGUUUAAACCCGAAUCUGAAUCUCGGAGUCCCGUCUUCGGCACCGACUUGGAGAUUCGUAUCUCAGCCGAAAGCGUCGUCGUCUUGGCCAUAAAAAUAU -(((..((.....(((((((.(((((......))))).(((((((.((((((((....)))....))))).)))))))......)))))))..))...)))................... ( -38.20) >DroSec_CAF1 13513 119 - 1 -GACAAGCAAACGUCGGCUUGGGUUUAAGCCCGAAUCUGAAUCUCGGAGUCCCGUCUUCGGCACCGACUUGGAGAUUCGUAUCUCAGCCGAAAGCGUCGUCGUCUUGGCCAUAAAAAUAU -(((..((.....(((((((((((....))))))....(((((((.((((((((....)))....))))).)))))))........)))))..))...)))................... ( -40.70) >DroSim_CAF1 14125 119 - 1 -GACAAGCAAACGUCGGCUUGGGUUUAAGCCCGAAUCUGAAUCUCGGAGUCCCGUCUUCGGCACCGACUUGGAGAUUCGUAUCUCAGCCGAAAGCGUCGUCGUCUUGGCCAUAAAAAUAU -(((..((.....(((((((((((....))))))....(((((((.((((((((....)))....))))).)))))))........)))))..))...)))................... ( -40.70) >DroEre_CAF1 14832 119 - 1 ACACAAGCAAAAGUCGGCU-UGGUUUAAGCCCGAAUCUGAAUCUCGGAGUACCGUCUUCGGCACCGACUUGGAGAUUCGUAUCUCAGCCGAAAGCGUCGUCGUCGUGGCCAUAAAAAUAU .(((.........((((((-.(((....))).((((.((((((((.((((.(.(((....).)).))))).)))))))).)).))))))))..(((....))).)))............. ( -33.60) >DroYak_CAF1 14844 119 - 1 -CACAUGCCAACGUCGGCUUUGGUUUAAGCCCGAAUCUGAAUCUCGGAGUCCCGUCUUCGGCAACGACUUGGAGAUUCGUAUCUCAGCCGAAAGCGUCGUCGUCUUGGCCAUAAAAAUAU -.....(((((.(.((((..((.(((..((..((((.((((((((.((((((((....)))....))))).)))))))).)).)).))..))).))..)))).))))))........... ( -38.50) >consensus _GACAAGCAAACGUCGGCUUUGGUUUAAGCCCGAAUCUGAAUCUCGGAGUCCCGUCUUCGGCACCGACUUGGAGAUUCGUAUCUCAGCCGAAAGCGUCGUCGUCUUGGCCAUAAAAAUAU ......((.....((((((..(((....))).((((.((((((((.((((((((....)))....))))).)))))))).)).))))))))..))((((......))))........... (-34.48 = -34.52 + 0.04)

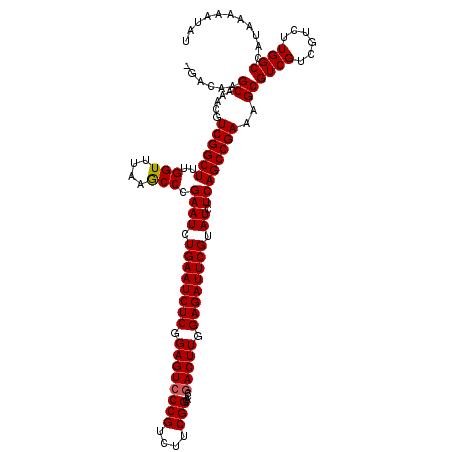
| Location | 359,443 – 359,546 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.90 |
| Mean single sequence MFE | -36.46 |
| Consensus MFE | -26.24 |
| Energy contribution | -26.60 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 359443 103 - 23771897 CACAGGCUUGAAGCUUCGUU-UUUGUAGCUUUAUUUGAGCCGCGCGCGGCUUCUGGCUCUCAAAA----------------GACAAGCAAACGUCGGCUGUGGUUUAAACCCGAAUCUGA ..(((..((((((((.((..-..)).)))))..((((((((((((.((((.....(((.((....----------------))..)))....)))))).))))))))))..)))..))). ( -31.30) >DroSec_CAF1 13593 103 - 1 CACAGGCUUGAAGCUUCGUU-UUUGUAGCUUUAUUUGAGCCGCGCGCGGCUUCUGGCUCUCAAAA----------------GACAAGCAAACGUCGGCUUGGGUUUAAGCCCGAAUCUGA ..(.((((((((.(....((-((((.((((......((((((....))))))..))))..)))))----------------).(((((........)))))).)))))))).)....... ( -33.50) >DroSim_CAF1 14205 103 - 1 CACAGGCUUGAAGCUUCGUU-UUUGUAGCUUUAUUUGAGCCGCGCGCGGCUUCUGGCUCUCAAAA----------------GACAAGCAAACGUCGGCUUGGGUUUAAGCCCGAAUCUGA ..(.((((((((.(....((-((((.((((......((((((....))))))..))))..)))))----------------).(((((........)))))).)))))))).)....... ( -33.50) >DroEre_CAF1 14912 119 - 1 CACGGGCUUGAAGCUGCGUUUUUUGUAGCUUUAUUUGAGCCGCGCGCGGCUUCUGGCUCUCAAAGCACAAAAACACAGAAACACAAGCAAAAGUCGGCU-UGGUUUAAGCCCGAAUCUGA ..(((((((((((((((((.....(..((((.....))))..)))))))))))(((((.....))).)).........((((.(((((........)))-)))))))))))))....... ( -44.10) >DroYak_CAF1 14924 106 - 1 CACGGGCUUGAAGCUCCGUUUUUUGUAGCUUUAUUUGAGCCGCGCGCGGCUUCUGGCUCUCAAGCCU--------------CACAUGCCAACGUCGGCUUUGGUUUAAGCCCGAAUCUGA ..(((((((((((((.((.....)).))))))((..((((((.(((.(((....((((....)))).--------------.....)))..)))))))))..))..)))))))....... ( -39.90) >consensus CACAGGCUUGAAGCUUCGUU_UUUGUAGCUUUAUUUGAGCCGCGCGCGGCUUCUGGCUCUCAAAA________________GACAAGCAAACGUCGGCUUUGGUUUAAGCCCGAAUCUGA ..((((((.((((((.(((.....(..((((.....))))..)))).)))))).)))....................................(((((((......))).))))..))). (-26.24 = -26.60 + 0.36)

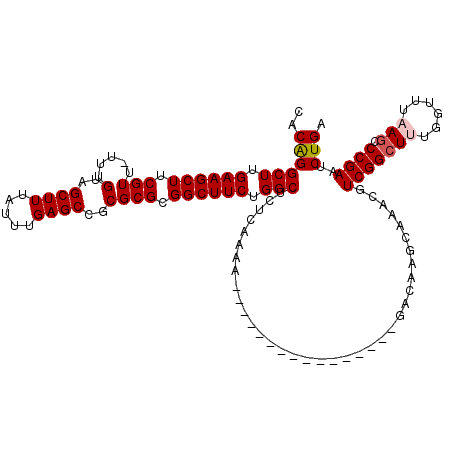
| Location | 359,482 – 359,586 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -26.96 |
| Consensus MFE | -22.84 |
| Energy contribution | -23.00 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 359482 104 + 23771897 ---------------UUUUGAGAGCCAGAAGCCGCGCGCGGCUCAAAUAAAGCUACAAA-AACGAAGCUUCAAGCCUGUGAUGAGCGCAUUAAAAUAAAUUUAAGAGCCCUCAAAAUGUU ---------------(((((((.(((....((.((.(((((((......(((((.(...-...).)))))..)))).)))....)))).(((((.....)))))).)).))))))).... ( -23.90) >DroSec_CAF1 13632 104 + 1 ---------------UUUUGAGAGCCAGAAGCCGCGCGCGGCUCAAAUAAAGCUACAAA-AACGAAGCUUCAAGCCUGUGAUGAGCGCAUUAAAAUAAAUUUAAGAGCCCUCAAAAUGUU ---------------(((((((.(((....((.((.(((((((......(((((.(...-...).)))))..)))).)))....)))).(((((.....)))))).)).))))))).... ( -23.90) >DroSim_CAF1 14244 104 + 1 ---------------UUUUGAGAGCCAGAAGCCGCGCGCGGCUCAAAUAAAGCUACAAA-AACGAAGCUUCAAGCCUGUGAUGAGCGCAUUAAAAUAAAUUUAAGAGCCCUCAAAAUGUU ---------------(((((((.(((....((.((.(((((((......(((((.(...-...).)))))..)))).)))....)))).(((((.....)))))).)).))))))).... ( -23.90) >DroEre_CAF1 14951 120 + 1 UUCUGUGUUUUUGUGCUUUGAGAGCCAGAAGCCGCGCGCGGCUCAAAUAAAGCUACAAAAAACGCAGCUUCAAGCCCGUGAUGAGCGCAUUAAAAUAAAUUUAAGAGCCCUCAAAAUGUU ..((((((((((((((((((.(((((....((...))..)))))...)))))).)).)))))))))).........(((..((((.((.(((((.....)))))..)).))))..))).. ( -33.50) >DroYak_CAF1 14963 106 + 1 -------------AGGCUUGAGAGCCAGAAGCCGCGCGCGGCUCAAAUAAAGCUACAAAAAACGGAGCUUCAAGCCCGUGAUGAGCGCAUUAAAAUAAAUUUAAGAGCCCUCA-AAUGUU -------------.((((((((((((....((...))..)))))......((((.(.......).)))))))))))(((..((((.((.(((((.....)))))..)).))))-.))).. ( -29.60) >consensus _______________UUUUGAGAGCCAGAAGCCGCGCGCGGCUCAAAUAAAGCUACAAA_AACGAAGCUUCAAGCCUGUGAUGAGCGCAUUAAAAUAAAUUUAAGAGCCCUCAAAAUGUU ................((((((.(((....((((((((.((((......(((((.(.......).)))))..)))))))).)).))...(((((.....)))))).)).))))))..... (-22.84 = -23.00 + 0.16)


| Location | 359,482 – 359,586 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -30.50 |
| Energy contribution | -30.30 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.69 |
| SVM RNA-class probability | 0.999938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 359482 104 - 23771897 AACAUUUUGAGGGCUCUUAAAUUUAUUUUAAUGCGCUCAUCACAGGCUUGAAGCUUCGUU-UUUGUAGCUUUAUUUGAGCCGCGCGCGGCUUCUGGCUCUCAAAA--------------- ....((((((((((..(((((.....)))))...........(((...(((((((.((..-..)).)))))))...((((((....)))))))))))))))))))--------------- ( -31.20) >DroSec_CAF1 13632 104 - 1 AACAUUUUGAGGGCUCUUAAAUUUAUUUUAAUGCGCUCAUCACAGGCUUGAAGCUUCGUU-UUUGUAGCUUUAUUUGAGCCGCGCGCGGCUUCUGGCUCUCAAAA--------------- ....((((((((((..(((((.....)))))...........(((...(((((((.((..-..)).)))))))...((((((....)))))))))))))))))))--------------- ( -31.20) >DroSim_CAF1 14244 104 - 1 AACAUUUUGAGGGCUCUUAAAUUUAUUUUAAUGCGCUCAUCACAGGCUUGAAGCUUCGUU-UUUGUAGCUUUAUUUGAGCCGCGCGCGGCUUCUGGCUCUCAAAA--------------- ....((((((((((..(((((.....)))))...........(((...(((((((.((..-..)).)))))))...((((((....)))))))))))))))))))--------------- ( -31.20) >DroEre_CAF1 14951 120 - 1 AACAUUUUGAGGGCUCUUAAAUUUAUUUUAAUGCGCUCAUCACGGGCUUGAAGCUGCGUUUUUUGUAGCUUUAUUUGAGCCGCGCGCGGCUUCUGGCUCUCAAAGCACAAAAACACAGAA ....(((((((((((...................((((.....))))..((((((((((.....(..((((.....))))..))))))))))).)))))))))))............... ( -39.20) >DroYak_CAF1 14963 106 - 1 AACAUU-UGAGGGCUCUUAAAUUUAUUUUAAUGCGCUCAUCACGGGCUUGAAGCUCCGUUUUUUGUAGCUUUAUUUGAGCCGCGCGCGGCUUCUGGCUCUCAAGCCU------------- ......-((((.((..(((((.....))))).)).))))....((((((((((((((((....((..((((.....))))..)).)))).....)))).))))))))------------- ( -34.10) >consensus AACAUUUUGAGGGCUCUUAAAUUUAUUUUAAUGCGCUCAUCACAGGCUUGAAGCUUCGUU_UUUGUAGCUUUAUUUGAGCCGCGCGCGGCUUCUGGCUCUCAAAA_______________ .....(((((((((..(((((.....)))))...........(((...(((((((.((.....)).)))))))...((((((....))))))))))))))))))................ (-30.50 = -30.30 + -0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:33 2006