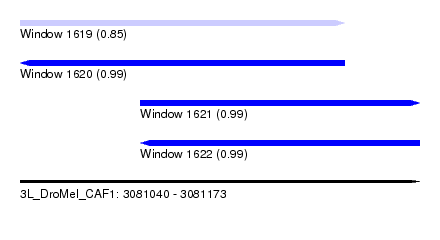
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,081,040 – 3,081,173 |
| Length | 133 |
| Max. P | 0.992884 |

| Location | 3,081,040 – 3,081,148 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 84.00 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -29.90 |
| Energy contribution | -29.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3081040 108 + 23771897 UUAUAUAAGAGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGUUU-UUUGCGUUUUUU--UCUUU----AUAC .((((.(((((((((....((((.......))))........(((((.((((.((.((((((.......))))).).)).))))...-)))))..)))))--)))))----))). ( -33.40) >DroVir_CAF1 30448 104 + 1 UUACAUAAGUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGGUA-A-ACC-UUUUU----GUUU----GUAC .((((.....(((((((..((((.......))))...........((((....))))(((((.......))))))))))))..((..-.-.))-.....----...)----))). ( -32.10) >DroGri_CAF1 25306 105 + 1 UUAUAUAAGUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUGAAAUUA-UCU-UUUUU----UUUU----CUGU ..........(((((((..((((.......))))...........((((....))))(((((.......)))))))))))).((((..(-...-...).----.)))----)... ( -30.10) >DroEre_CAF1 18164 109 + 1 UUAUAUAAGUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGUUU-UUUGU-UUUGUUUCGCCUU----UUCU ..........((.((....((((.......))))....(((.(((((.((((.((.((((((.......))))).).)).))))...-)))))-...))))).))..----.... ( -31.00) >DroMoj_CAF1 26744 109 + 1 UUAUAUAAGAGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGGUA-A-AGC-UUUUUU---GCUUACUUUUAU ........(((((((((..((((.......))))...........((((....))))(((((.......))))))))))))))((((-(-.((-......---)))))))..... ( -37.10) >DroAna_CAF1 12970 94 + 1 UUAUAUAAGAGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUAAUUU-UUU--------U--UCU---------- .........((((((((..((((.......))))...........((((....))))(((((.......))))))))))))).....-...--------.--...---------- ( -31.30) >consensus UUAUAUAAGAGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGUUU_A_UGC_UUUUUU___CUUU____UUAU ..........(((((((..((((.......))))...........((((....))))(((((.......)))))))))))).................................. (-29.90 = -29.90 + 0.00)

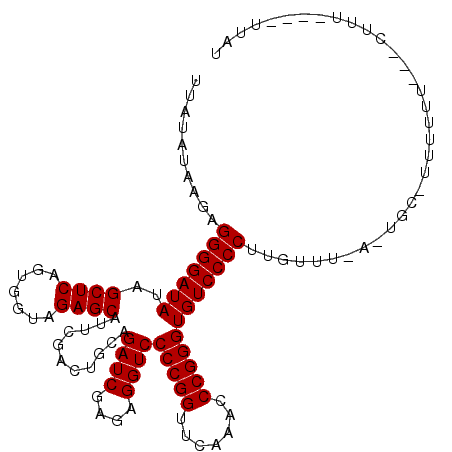
| Location | 3,081,040 – 3,081,148 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 84.00 |
| Mean single sequence MFE | -30.40 |
| Consensus MFE | -28.65 |
| Energy contribution | -28.65 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.990890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3081040 108 - 23771897 GUAU----AAAGA--AAAAAACGCAAA-AAACAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCUCUUAUAUAA ((((----((.((--............-((((..(((....)))..)))).....(((((....(((((.....)))))..((((.......))))....))))))))))))).. ( -29.60) >DroVir_CAF1 30448 104 - 1 GUAC----AAAC----AAAAA-GGU-U-UACCAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCACUUAUGUAA .(((----(...----.....-(((-(-((((..(((....))).)))..)))))(((((....(((((.....)))))..((((.......))))....))))).....)))). ( -33.90) >DroGri_CAF1 25306 105 - 1 ACAG----AAAA----AAAAA-AGA-UAAUUUCAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCACUUAUAUAA ....----....----.....-...-........(((((..(((((.......)))))......(((((.....)))))..((((.......))))....))))).......... ( -28.90) >DroEre_CAF1 18164 109 - 1 AGAA----AAGGCGAAACAAA-ACAAA-AAACAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCACUUAUAUAA ....----.............-.....-......(((((..(((((.......)))))......(((((.....)))))..((((.......))))....))))).......... ( -28.90) >DroMoj_CAF1 26744 109 - 1 AUAAAAGUAAGC---AAAAAA-GCU-U-UACCAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCUCUUAUAUAA ......((((((---......-)).-)-)))..((((((..(((((.......)))))......(((((.....)))))..((((.......))))....))))))......... ( -32.20) >DroAna_CAF1 12970 94 - 1 ----------AGA--A--------AAA-AAAUUAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCUCUUAUAUAA ----------...--.--------...-.....((((((..(((((.......)))))......(((((.....)))))..((((.......))))....))))))......... ( -28.90) >consensus AUAA____AAAG___AAAAAA_GCA_A_AAACAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCACUUAUAUAA ..................................(((((..(((((.......)))))......(((((.....)))))..((((.......))))....))))).......... (-28.65 = -28.65 + 0.00)

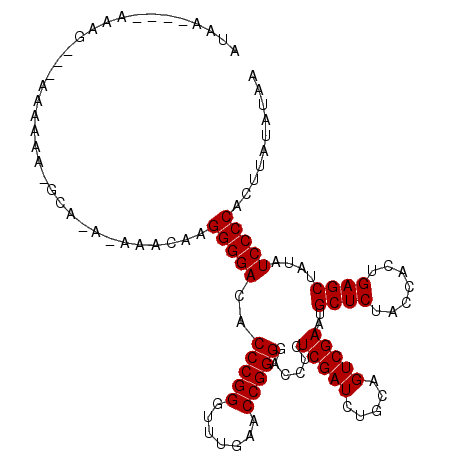
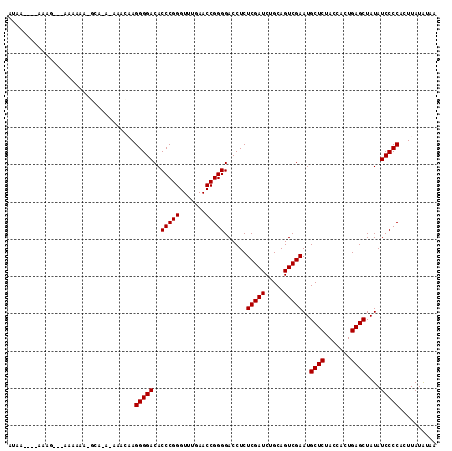
| Location | 3,081,080 – 3,081,173 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 71.38 |
| Mean single sequence MFE | -21.82 |
| Consensus MFE | -15.32 |
| Energy contribution | -15.65 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3081080 93 + 23771897 CUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGUUU-UUUGCGUUUUUU--UCUUU----AUACCCAAUUACUUGUUCAUGUUUAAUUU------- ..(((((.((((.((.((((((.......))))).).)).))))...-))))).......--.....----.............................------- ( -22.50) >DroVir_CAF1 30488 84 + 1 CUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGGUA-A-ACC-UUUUU----GUUU----GUACUUAAGCA-----UAAUAAUUAAUUU------- ......((((((.((.((((((.......))))).).)).)))))).-.-...-....(----((((----(....))))))-----.............------- ( -21.80) >DroGri_CAF1 25346 90 + 1 CUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUGAAAUUA-UCU-UUUUU----UUUU----CUGUUUAAGAA-----UCCUGUU--UGUAAUAAUCU .(((((((.(((.((.((((((.......))))).).)).)).......-...-.....----..((----((.....))))-----..).)))--))))....... ( -19.80) >DroSec_CAF1 13993 80 + 1 CUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGCUU-U-UGC-CUUUUU--UCUUU----AUACCC-----------AAUGUUUAAUUU------- ..(((((.((((.((.((((((.......))))).).)).))))..)-)-)))-......--.....----......-----------............------- ( -21.50) >DroYak_CAF1 14804 94 + 1 CUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGUUU-UUUGC-CUUCUUUCUCUUU----UUGCCCAAUUACUUGUUCAUGUUCAAUUU------- ..(((((.((((.((.((((((.......))))).).)).))))...-)))))-.............----.((..(((....)))..))..........------- ( -21.70) >DroMoj_CAF1 26784 88 + 1 CUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGGUA-A-AGC-UUUUUU---GCUUACUUUUAUUUUCAGA-----U-UUAUUUAAGUA------- (((...((((((.((.((((((.......))))).).)).)))))).-(-(((-......---))))...........))).-----.-...........------- ( -23.60) >consensus CUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGGUU_A_UGC_UUUUUU___CUUU____AUACCCAAGUA_____UCAUGUUUAAUUU_______ ........((((.((.((((((.......))))).).)).))))............................................................... (-15.32 = -15.65 + 0.33)
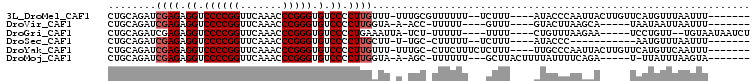
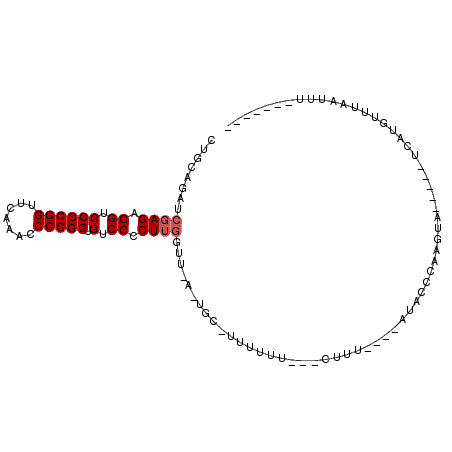
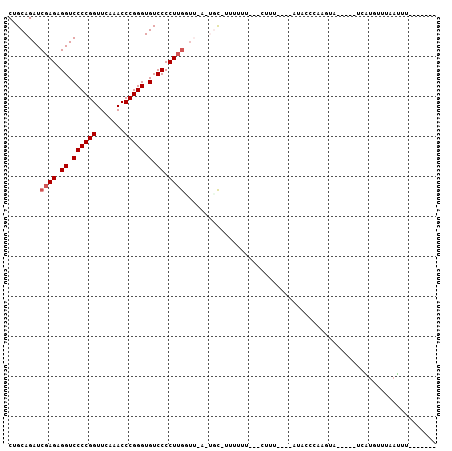
| Location | 3,081,080 – 3,081,173 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 71.38 |
| Mean single sequence MFE | -21.48 |
| Consensus MFE | -16.84 |
| Energy contribution | -16.70 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.990871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3081080 93 - 23771897 -------AAAUUAAACAUGAACAAGUAAUUGGGUAU----AAAGA--AAAAAACGCAAA-AAACAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAG -------.........(((..(((....)))..)))----.....--.......(((..-...(.(((((...(((((.......)))))..))))).)...))).. ( -22.70) >DroVir_CAF1 30488 84 - 1 -------AAAUUAAUUAUUA-----UGCUUAAGUAC----AAAC----AAAAA-GGU-U-UACCAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAG -------.............-----(((........----....----.....-((.-.-..)).(((((...(((((.......)))))..)))))......))). ( -20.70) >DroGri_CAF1 25346 90 - 1 AGAUUAUUACA--AACAGGA-----UUCUUAAACAG----AAAA----AAAAA-AGA-UAAUUUCAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAG ...........--..((((.-----((((.....))----))..----.....-...-.....(((((((...(((((.......)))))..))))).))))))... ( -20.70) >DroSec_CAF1 13993 80 - 1 -------AAAUUAAACAUU-----------GGGUAU----AAAGA--AAAAAG-GCA-A-AAGCAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAG -------............-----------......----.....--......-(((-.-...(.(((((...(((((.......)))))..))))).)...))).. ( -20.40) >DroYak_CAF1 14804 94 - 1 -------AAAUUGAACAUGAACAAGUAAUUGGGCAA----AAAGAGAAAGAAG-GCAAA-AAACAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAG -------..........((..(((....)))..)).----.............-(((..-...(.(((((...(((((.......)))))..))))).)...))).. ( -23.30) >DroMoj_CAF1 26784 88 - 1 -------UACUUAAAUAA-A-----UCUGAAAAUAAAAGUAAGC---AAAAAA-GCU-U-UACCAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAG -------...........-.-----.(((.........((((((---......-)).-)-)))(.(((((...(((((.......)))))..))))).).....))) ( -21.10) >consensus _______AAAUUAAACAUGA_____UAAUUAAGUAA____AAAG___AAAAAA_GCA_A_AAACAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAG ...............................................................(.(((((...(((((.......)))))..))))).)........ (-16.84 = -16.70 + -0.14)

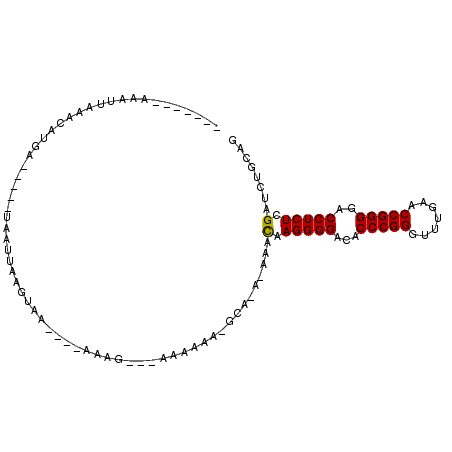
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:44 2006