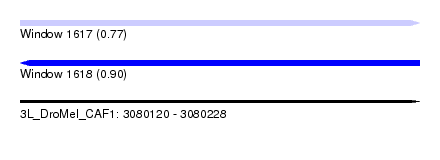
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,080,120 – 3,080,228 |
| Length | 108 |
| Max. P | 0.900615 |

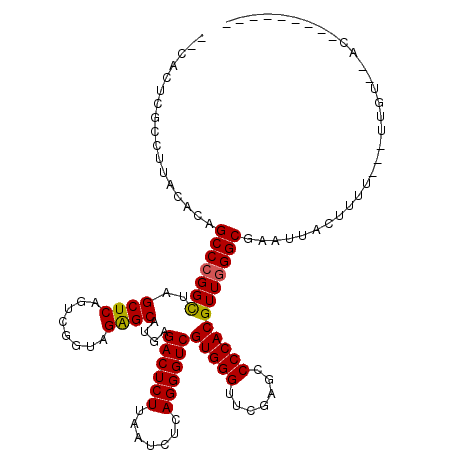
| Location | 3,080,120 – 3,080,228 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.61 |
| Mean single sequence MFE | -34.88 |
| Consensus MFE | -30.78 |
| Energy contribution | -30.66 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3080120 108 + 23771897 --CACUUGCCUGACACAGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAUUAUUUUU---UAGUUCACA-----C-- --.....(((..((...((((.....((((........)))).(((((.......))))))))).(((((.......)))))))..)))((((((.....---))))))...-----.-- ( -39.40) >DroSec_CAF1 13094 102 + 1 GCCCCUUCAAUAACCCGGCCCGGUUAGCUCAGUGGGUAGGGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUAGGCGUUUUACUUUU---UU--------------- (((.........((((.((((.....(((.....))).)))).(((((.......)))))))))((((((.......))))))...)))...........---..--------------- ( -32.50) >DroSim_CAF1 13050 100 + 1 --CGCUCGCCUCACAUAGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAUUAUAUUU---UA--------------- --...((((((.((...((((.....((((........)))).(((((.......))))))))).(((((.......))))))).)))))).........---..--------------- ( -35.70) >DroEre_CAF1 17202 107 + 1 --CGCUCAUUCUUCACAGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAUG--CUUUU---UUGUUUACU------UU --((((((.........((((.....((((........)))).(((((.......)))))))))((((((.......)))))).))))))...--.....---.........------.. ( -33.20) >DroYak_CAF1 13739 116 + 1 --CACUAUUCUUGCACAGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAA--CUUUUUUUUUGUAUACUUUACGUUU --...............(((((((..((((........))))....((((((.......))))))(((((.......))))))))))))....--..........(((.....))).... ( -33.60) >consensus __CACUCGCCUUACACAGCCCGGCUAGCUCAGUCGGUAGAGCAUGAGACUCUUAAUCUCAGGGUCGUGGGUUCGAGCCCCACGUUGGGCGAAUUACUUUU___UUGU__AC_________ .................(((((((..((((........))))....((((((.......))))))(((((.......))))))))))))............................... (-30.78 = -30.66 + -0.12)

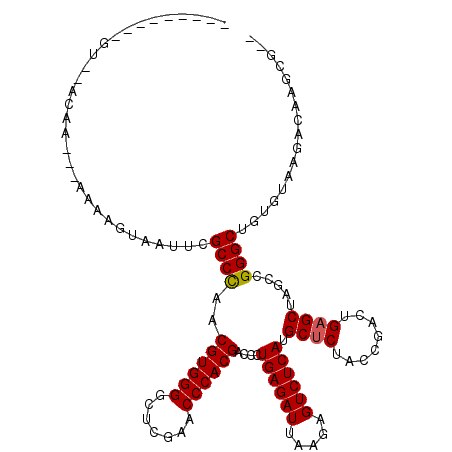
| Location | 3,080,120 – 3,080,228 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.61 |
| Mean single sequence MFE | -33.57 |
| Consensus MFE | -29.44 |
| Energy contribution | -29.48 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3080120 108 - 23771897 --G-----UGUGAACUA---AAAAAUAAUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCUGUGUCAGGCAAGUG-- --(-----(.(((....---...........((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))....))).)).....-- ( -32.81) >DroSec_CAF1 13094 102 - 1 ---------------AA---AAAAGUAAAACGCCUAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCCCUACCCACUGAGCUAACCGGGCCGGGUUAUUGAAGGGGC ---------------..---...........((((..((((((.......))))))((((((((((.....)))))).((((.................)))).))))........)))) ( -32.93) >DroSim_CAF1 13050 100 - 1 ---------------UA---AAAUAUAAUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCUAUGUGAGGCGAGCG-- ---------------..---........((((((((.((((((.......)))))).(((((((((.....)))))).((((........)))).....))).....)).))))))..-- ( -36.80) >DroEre_CAF1 17202 107 - 1 AA------AGUAAACAA---AAAAG--CAUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCUGUGAAGAAUGAGCG-- ..------.........---.....--(((.((((..((((((.......))))))....((((((.....)))))).((((........)))).....)))).)))...........-- ( -33.20) >DroYak_CAF1 13739 116 - 1 AAACGUAAAGUAUACAAAAAAAAAG--UUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCUGUGCAAGAAUAGUG-- ....(((.....))).........(--(...((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))...))..........-- ( -32.10) >consensus _________GU__ACAA___AAAAGUAAUUCGCCCAACGUGGGGCUCGAACCCACGACCCUGAGAUUAAGAGUCUCAUGCUCUACCGACUGAGCUAGCCGGGCUGUGUAAGACAAGCG__ ...............................((((..((((((.......))))))....((((((.....)))))).((((........)))).....))))................. (-29.44 = -29.48 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:40 2006