
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,079,888 – 3,079,990 |
| Length | 102 |
| Max. P | 0.990053 |

| Location | 3,079,888 – 3,079,990 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 86.16 |
| Mean single sequence MFE | -32.93 |
| Consensus MFE | -29.80 |
| Energy contribution | -29.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
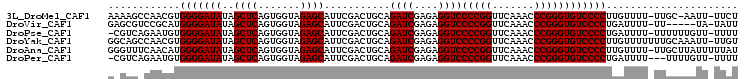
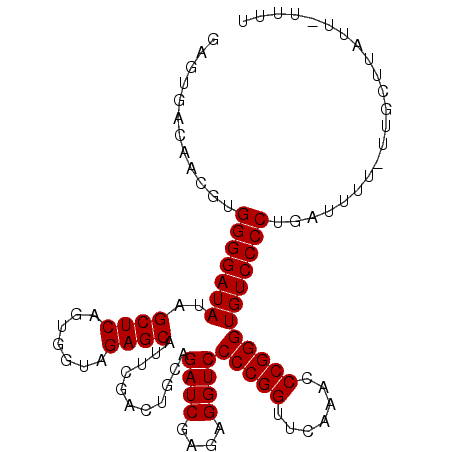
>3L_DroMel_CAF1 3079888 102 + 23771897 AAAAGCCAACGUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGUUUU-UUGC-AAUU-UUCU ....((.((((.(((((((..((((.......))))...........((((....))))(((((.......)))))))))))).))))..-..))-....-.... ( -33.30) >DroVir_CAF1 29911 98 + 1 GAGCGUCCGCAUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUGAUUUU-UU-----UA-UAUU ..((....))..(((((((..((((.......))))...........((((....))))(((((.......)))))))))))).......-..-----..-.... ( -31.50) >DroPse_CAF1 15379 102 + 1 -CGUCAGAAUGUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUGAUUUU-UUUUUUGUU-UUUU -....(((((..(((((((..((((.......))))...........((((....))))(((((.......))))))))))))..)))))-.........-.... ( -30.90) >DroYak_CAF1 13505 104 + 1 GGCAGCCAACGUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGUUUUUUUGCAAAUU-UUGU .((((..((((.(((((((..((((.......))))...........((((....))))(((((.......)))))))))))).))))...)))).....-.... ( -36.10) >DroAna_CAF1 12279 104 + 1 GGGUUUCAACAUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGUUUU-UUGCUUAUUUUUAU ((((((.(((.(((((((...((((.......)))).((((((....).)))))..)))))))))).))))))(((....))).......-.............. ( -34.90) >DroPer_CAF1 15545 100 + 1 -CGUCAGAAUGUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUGAUUUU---UUUUGUU-UUUU -....(((((..(((((((..((((.......))))...........((((....))))(((((.......))))))))))))..)))))---.......-.... ( -30.90) >consensus GAGUGACAACGUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUGAUUUU_UUGCUUAUU_UUUU ............(((((((..((((.......))))...........((((....))))(((((.......))))))))))))...................... (-29.80 = -29.80 + 0.00)
| Location | 3,079,888 – 3,079,990 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 86.16 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -28.90 |
| Energy contribution | -28.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.990053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
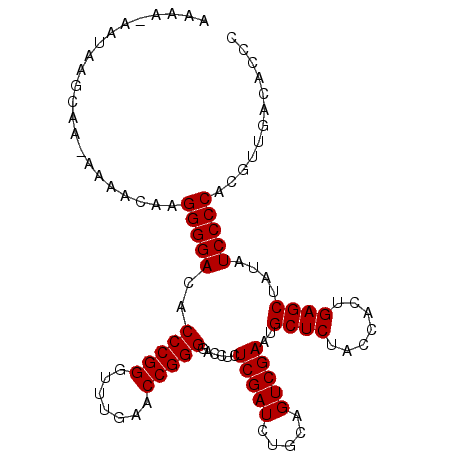
>3L_DroMel_CAF1 3079888 102 - 23771897 AGAA-AAUU-GCAA-AAAACAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCACGUUGGCUUUU ....-....-((..-..(((..(((((..(((((.......)))))......(((((.....)))))..((((.......))))....)))))..))).)).... ( -31.30) >DroVir_CAF1 29911 98 - 1 AAUA-UA-----AA-AAAAUCAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCAUGCGGACGCUC ....-..-----..-....(((((((...(((((.......)))))..))))).))(((((((..((..((((.......))))....))..).))))))..... ( -30.80) >DroPse_CAF1 15379 102 - 1 AAAA-AACAAAAAA-AAAAUCAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCACAUUCUGACG- ....-.........-.......(((((..(((((.......)))))......(((((.....)))))..((((.......))))....)))))...........- ( -28.90) >DroYak_CAF1 13505 104 - 1 ACAA-AAUUUGCAAAAAAACAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCACGUUGGCUGCC ....-.....(((....(((..(((((..(((((.......)))))......(((((.....)))))..((((.......))))....)))))..)))...))). ( -31.80) >DroAna_CAF1 12279 104 - 1 AUAAAAAUAAGCAA-AAAACAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCAUGUUGAAACCC ..............-.......(((....)))((((((.(((.(((((....(((((.....)))))..((((.......))))....)))))..))).)))))) ( -34.30) >DroPer_CAF1 15545 100 - 1 AAAA-AACAAAA---AAAAUCAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCACAUUCUGACG- ....-.......---.......(((((..(((((.......)))))......(((((.....)))))..((((.......))))....)))))...........- ( -28.90) >consensus AAAA_AAUAAGCAA_AAAACAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCACGUUGACACCC ......................(((((..(((((.......)))))......(((((.....)))))..((((.......))))....)))))............ (-28.90 = -28.90 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:38 2006