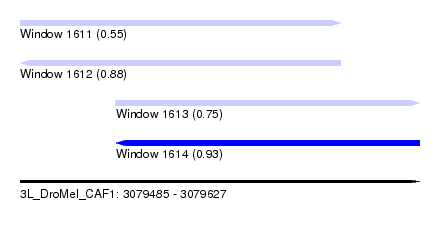
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,079,485 – 3,079,627 |
| Length | 142 |
| Max. P | 0.926672 |

| Location | 3,079,485 – 3,079,599 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.20 |
| Mean single sequence MFE | -40.25 |
| Consensus MFE | -29.80 |
| Energy contribution | -29.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
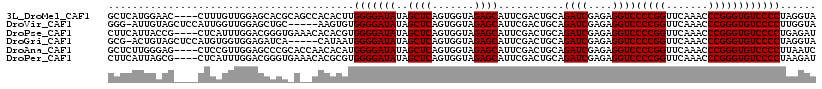
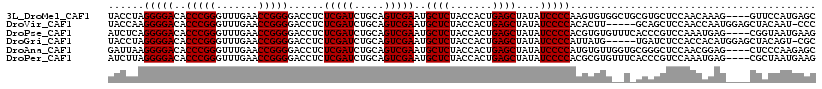
>3L_DroMel_CAF1 3079485 114 + 23771897 GCUCAUGGAAC----CUUUGUUGGAGCACGCAGCCACACUUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUAGGUA ((((....(((----....))).))))..........((((((((.....((((.......))))....(((....((((....))))(((((.......))))).))))))))))). ( -37.80) >DroVir_CAF1 29421 112 + 1 GGG-AUUGUAGCUCCAUUGGUUGGAGCUGC-----AAGUGUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGGUA ...-.(((((((((((.....)))))))))-----))....(((((((..((((.......))))...........((((....))))(((((.......))))))))))))...... ( -47.80) >DroPse_CAF1 15026 114 + 1 CUUCAUUACCG----CUCAUUUGGACGGGUGAAACACACGUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUGAGAU .(((((..(((----..(....)..)))))))).....((.(((((((..((((.......))))...........((((....))))(((((.......)))))))))))))).... ( -37.20) >DroGri_CAF1 24351 112 + 1 GCG-ACUGUAGCUCCAUGUGGUGGAGAUCA-----CAUAAUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUAGGUA .((-(((((((((((((...(((.....))-----)...)))))).....((((.......))))......)))))).)))((.((.((((((.......))))).).)).))..... ( -40.10) >DroAna_CAF1 11842 114 + 1 GCUCUUGGGAG----CUCCGUUGGAGCCCGCACCAACACAUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUAAUC ((((....)))----)...(((((.(....).)))))....(((((((..((((.......))))...........((((....))))(((((.......))))))))))))...... ( -42.00) >DroPer_CAF1 15198 114 + 1 CUUCAUUAGCG----CUCAUUUGGACGGGUGAAACACGCGUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUAAGAU ........(((----(((((((....)))))).....))))(((((((..((((.......))))...........((((....))))(((((.......))))))))))))...... ( -36.60) >consensus GCUCAUUGGAG____CUCAGUUGGAGCGCG_A__CACACGUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUAAGUA .........................................(((((((..((((.......))))...........((((....))))(((((.......))))))))))))...... (-29.80 = -29.80 + 0.00)
| Location | 3,079,485 – 3,079,599 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.20 |
| Mean single sequence MFE | -38.38 |
| Consensus MFE | -28.90 |
| Energy contribution | -28.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3079485 114 - 23771897 UACCUAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCAAGUGUGGCUGCGUGCUCCAACAAAG----GUUCCAUGAGC .((((.(((....)))))))..((((((((((....(((((.....)))))..((((.......))))....)))))...((..((.....))..)).....)----))))....... ( -38.10) >DroVir_CAF1 29421 112 - 1 UACCAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCACACUU-----GCAGCUCCAACCAAUGGAGCUACAAU-CCC ......(((((..(((((.......)))))......(((((.....)))))..((((.......))))....)))))....((-----(.(((((((.....))))))).))).-... ( -41.10) >DroPse_CAF1 15026 114 - 1 AUCUCAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCACGUGUGUUUCACCCGUCCAAAUGAG----CGGUAAUGAAG ...(((.(..((((.(((((....)))(((((....(((((.....)))))..((((.......))))....))))).)).))))..)..(((..(....)..----)))...))).. ( -37.20) >DroGri_CAF1 24351 112 - 1 UACCUAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCAUUAUG-----UGAUCUCCACCACAUGGAGCUACAGU-CGC ......(((((..(((((.......)))))......(((((.....)))))..((((.......))))....))))).....(-----(((((((((.....))))).....))-))) ( -36.50) >DroAna_CAF1 11842 114 - 1 GAUUAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCAUGUGUUGGUGCGGGCUCCAACGGAG----CUCCCAAGAGC (((..(((((...(((((.......)))))..)))))..))).(((.......((((.......)))).......(((.....))))))(((((((...))))----)))........ ( -42.10) >DroPer_CAF1 15198 114 - 1 AUCUUAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCACGCGUGUUUCACCCGUCCAAAUGAG----CGCUAAUGAAG .(((((((..((((.(((((....)))(((((....(((((.....)))))..((((.......))))....))))).)).))))..)...((..(....)..----)))))).)).. ( -35.30) >consensus UACCUAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCACGUGUG__U_ACAGCUCCAACAGAG____CUACAAUGAGC ......(((((..(((((.......)))))......(((((.....)))))..((((.......))))....)))))......................................... (-28.90 = -28.90 + 0.00)
| Location | 3,079,519 – 3,079,627 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.15 |
| Mean single sequence MFE | -33.69 |
| Consensus MFE | -29.80 |
| Energy contribution | -29.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
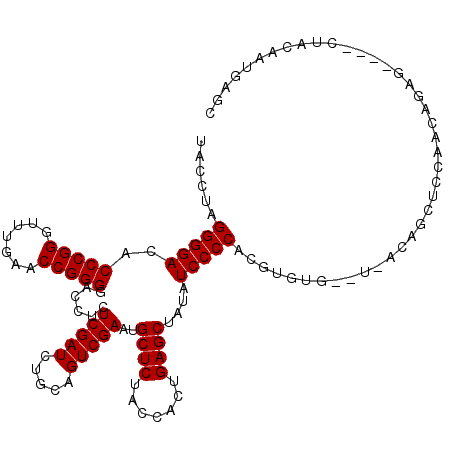
>3L_DroMel_CAF1 3079519 108 + 23771897 CUUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUAGGUAAACUUUU-UUC----CCAUCUGCCU-----AAUAUUAU ...(((((((..((((.......))))...........((((....))))(((((.......))))))))))))((((((.......-...----.....)))))-----)....... ( -33.96) >DroVir_CAF1 29453 112 + 1 UGUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGGUAAACUUUU-UGU----GUUGCCUUUUGCUUUAAUUUUU- ...(((((((..((((.......))))...........((((....))))(((((.......))))))))))))..(((((((....-.))----.)))))................- ( -34.40) >DroPse_CAF1 15060 111 + 1 CGUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUGAGAUUACUUUUUUUGUUUUUCAUCUGUAA-----AGGAUU-- ...(((((((..((((.......))))...........((((....))))(((((.......))))))))))))(((((..((.......)).)))))(((....-----.)))..-- ( -32.10) >DroGri_CAF1 24383 102 + 1 AAUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUAGGUAAACCUUU-UUG----A-U---------GUCAAUAUUC- ...(((((((..((((.......))))...........((((....))))(((((.......)))))))))))).(((....)))..-...----.-.---------..........- ( -31.30) >DroMoj_CAF1 25722 112 + 1 CAUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGGUAAACUUUU-UUG----CCAGCUUCCCCCAUUAAUUUUU- .((((((....((((....(((((((((......)))..((((((.((.((((((.......))))).).)).)))))).......)-)))----))))))..))))))........- ( -39.30) >DroAna_CAF1 11876 106 + 1 CAUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUAAUCA-CUUUU-UUG----UUUCAUAC-A-----AGUUUUAA ...(((((((..((((.......))))...........((((....))))(((((.......)))))))))))).......-....(-(((----(.....))-)-----))...... ( -31.10) >consensus CAUGGGGAUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAACCCGGGUGUCCCCUUGGUAAACUUUU_UUG____CCAGCUGC_A_____AAUAUUA_ ...(((((((..((((.......))))...........((((....))))(((((.......))))))))))))............................................ (-29.80 = -29.80 + 0.00)

| Location | 3,079,519 – 3,079,627 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.15 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -28.90 |
| Energy contribution | -28.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
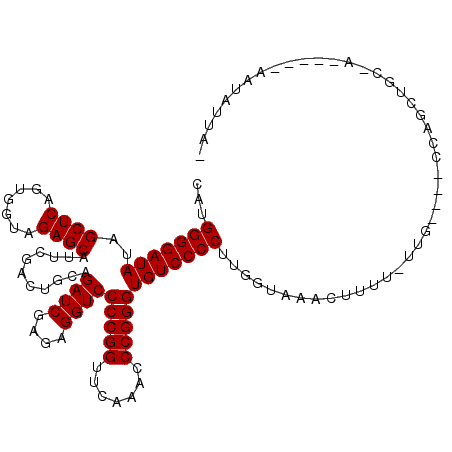
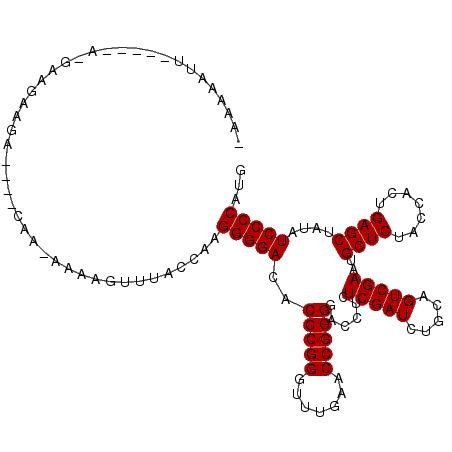
>3L_DroMel_CAF1 3079519 108 - 23771897 AUAAUAUU-----AGGCAGAUGG----GAA-AAAAGUUUACCUAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCAAG .......(-----(((.((((..----...-....)))).))))(((((..(((((.......)))))......(((((.....)))))..((((.......))))....)))))... ( -32.70) >DroVir_CAF1 29453 112 - 1 -AAAAAUUAAAGCAAAAGGCAAC----ACA-AAAAGUUUACCAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCACA -................(....)----...-.............(((((..(((((.......)))))......(((((.....)))))..((((.......))))....)))))... ( -30.10) >DroPse_CAF1 15060 111 - 1 --AAUCCU-----UUACAGAUGAAAAACAAAAAAAGUAAUCUCAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCACG --......-----...............................(((((..(((((.......)))))......(((((.....)))))..((((.......))))....)))))... ( -28.90) >DroGri_CAF1 24383 102 - 1 -GAAUAUUGAC---------A-U----CAA-AAAGGUUUACCUAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCAUU -.....(((..---------.-.----)))-...(((((((((.(((....)))))))..)))))(((((....(((((.....)))))..((((.......))))....)))))... ( -33.30) >DroMoj_CAF1 25722 112 - 1 -AAAAAUUAAUGGGGGAAGCUGG----CAA-AAAAGUUUACCAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCAUG -........((((((((((((..----...-...))))).....((((...(((((.......)))))..))))(((((.....)))))..((((.......))))....))))))). ( -37.50) >DroAna_CAF1 11876 106 - 1 UUAAAACU-----U-GUAUGAAA----CAA-AAAAG-UGAUUAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCAUG ((((.(((-----(-........----...-..)))-)..))))(((((..(((((.......)))))......(((((.....)))))..((((.......))))....)))))... ( -29.62) >consensus _AAAAAUU_____A_GAAGAAGA____CAA_AAAAGUUUACCAAGGGGACACCCGGGUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUAUCCCCAUG ............................................(((((..(((((.......)))))......(((((.....)))))..((((.......))))....)))))... (-28.90 = -28.90 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:36 2006