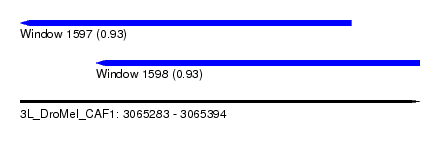
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,065,283 – 3,065,394 |
| Length | 111 |
| Max. P | 0.930771 |

| Location | 3,065,283 – 3,065,375 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 80.55 |
| Mean single sequence MFE | -30.17 |
| Consensus MFE | -16.91 |
| Energy contribution | -16.80 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3065283 92 - 23771897 GCA----GGCCAAACAAUAACAAA--AACCGCGCUGUUACGUCACGUUAAGUGAAUAUGUAGAUGUGGCAGAGCGGU----CUUAAGGCCUACCAUUUACCG ..(----((((.............--.(((((.((((((((((((((.........)))).)))))))))).)))))----.....)))))........... ( -30.41) >DroPse_CAF1 32024 88 - 1 GCAGGUAAACCAAACAAUAACAAA--AAGCCCGCUGUUACGUCACGUAA----AAUGCGUUGAUGUGGCAGCACGGGCAACA-CGUCG-------UUUACCG ...(((((((..............--......(((((((((((((((..----...))).))))))))))))((((....).-))).)-------)))))). ( -33.80) >DroSec_CAF1 26048 94 - 1 GCA----GGCCAAACAAUAACAAA--AACCCCGCUGUUACGUCACGUUUAGUGAAUAUGUAGAUGUGGCAGAGCGGU--ACGUUACGGCCUACCAUUUACCG ..(----((((......((((...--.(((.(.((((((((((((((.........)))).)))))))))).).)))--..)))).)))))........... ( -26.30) >DroSim_CAF1 26019 94 - 1 GCA----GGCCAAACAAUAACAAA--AACCGCGCUGUUACGUCACGUUUAGUGAAUAUGUAGAUGUGGCAGAGCGGU--ACGUUACGGCCUACCAUUUACCG ..(----((((......((((...--.(((((.((((((((((((((.........)))).)))))))))).)))))--..)))).)))))........... ( -32.60) >DroYak_CAF1 26241 95 - 1 GCA----GGCCAAACAAUAACAAAAAAACCGCGCUGUUACGUCACGUUAAGUGAAUAUGUAGAUGUGGCAGAGCGGU--ACG-GAUGGUCUACCAUUUACCG ...----((..................(((((.((((((((((((((.........)))).)))))))))).)))))--..(-(((((....)))))).)). ( -31.80) >DroAna_CAF1 27991 94 - 1 GCA----GGCUAAACAAUAACAAA--AAGUACGCUGUUACGUCACGU-AAGUUCAUAUGUAGAUGUGGCAGAGCGGGUGUCG-CUUAGUUUACCAUUUACCG ...----((.(((((.........--.......((((((((((((((-(......))))).))))))))))((((.....))-))..)))))))........ ( -26.10) >consensus GCA____GGCCAAACAAUAACAAA__AACCGCGCUGUUACGUCACGUUAAGUGAAUAUGUAGAUGUGGCAGAGCGGU__ACG_UAUGGCCUACCAUUUACCG .......((((......................((((((((((((((.........)))).))))))))))...(.....).....))))............ (-16.91 = -16.80 + -0.11)

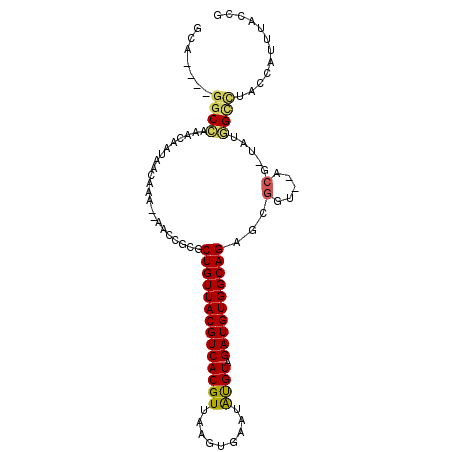
| Location | 3,065,304 – 3,065,394 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.37 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -15.66 |
| Energy contribution | -16.08 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3065304 90 - 23771897 --A--------UAGUCACACGUU----UGGGGAGCA----GGCCAAACAAUAACAAA--AACCGCGCUGUUACGUCACGUUAAGUGAAUAUGUAGAUGUGGCAGAGCGGU --.--------.........(((----(((......----..)))))).........--.(((((.((((((((((((((.........)))).)))))))))).))))) ( -31.70) >DroPse_CAF1 32041 87 - 1 --A--------UGCUGGCAC-------UGAGAGGCAGGUAAACCAAACAAUAACAAA--AAGCCCGCUGUUACGUCACGUAA----AAUGCGUUGAUGUGGCAGCACGGG --.--------....(((.(-------(....))..((....)).............--..))).(((((((((((((((..----...))).))))))))))))..... ( -28.30) >DroSec_CAF1 26071 92 - 1 AGA--------UAGUCACACGUU----UGGGGAGCA----GGCCAAACAAUAACAAA--AACCCCGCUGUUACGUCACGUUUAGUGAAUAUGUAGAUGUGGCAGAGCGGU ...--------.........(((----(((......----..)))))).........--.(((.(.((((((((((((((.........)))).)))))))))).).))) ( -25.40) >DroSim_CAF1 26042 92 - 1 AGA--------UAGUCACACGUU----UGGGGAGCA----GGCCAAACAAUAACAAA--AACCGCGCUGUUACGUCACGUUUAGUGAAUAUGUAGAUGUGGCAGAGCGGU ...--------.........(((----(((......----..)))))).........--.(((((.((((((((((((((.........)))).)))))))))).))))) ( -31.70) >DroYak_CAF1 26263 102 - 1 AGAUAGUCAGAUAGUCACACGUU----UGGGGAGCA----GGCCAAACAAUAACAAAAAAACCGCGCUGUUACGUCACGUUAAGUGAAUAUGUAGAUGUGGCAGAGCGGU ....................(((----(((......----..))))))............(((((.((((((((((((((.........)))).)))))))))).))))) ( -31.70) >DroAna_CAF1 28015 91 - 1 -----------UACU-ACAUAUUGUGGGGGGGAGCA----GGCUAAACAAUAACAAA--AAGUACGCUGUUACGUCACGU-AAGUUCAUAUGUAGAUGUGGCAGAGCGGG -----------....-...((((((..((.......----..))..)))))).....--.....((((((((((((((((-(......))))).))))))))..)))).. ( -21.90) >consensus __A________UAGUCACACGUU____UGGGGAGCA____GGCCAAACAAUAACAAA__AACCGCGCUGUUACGUCACGUUAAGUGAAUAUGUAGAUGUGGCAGAGCGGU ............................................................(((.(.((((((((((((((.........)))).)))))))))).).))) (-15.66 = -16.08 + 0.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:21 2006