| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,064,379 – 3,064,490 |
| Length | 111 |
| Max. P | 0.680664 |

| Location | 3,064,379 – 3,064,490 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 87.44 |
| Mean single sequence MFE | -28.85 |
| Consensus MFE | -19.17 |
| Energy contribution | -19.61 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.680664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3064379 111 + 23771897 GAUUAGCUGGCCCAAACUACAAUCGUGGGAGUAUUAAACUAUACGCUUGAUAG-GCUAAUUAAUAGAAGAAUUGAGCAGUUGGGUAACAUUAUGGCUAGUCUUCGUAUUUAA ((((((((.((((((.((((....))))..((((......))))(((..((..-.(((.....)))....))..)))..))))))........))))))))........... ( -28.60) >DroSec_CAF1 25132 111 + 1 GAUUAGCUGGCCCAAACUACAAUCGUGGGUGUAUUAAACUAUACGCUUGAUAG-GCUAAUUAAUAGAAAAAUUGAGCCGUUGGGUAACAUUAUGGCUAGCCUUCGUAUUUAA ((...(((((((....((((....))))(((((......)))))(((..((.(-(((((((........)))).))))))..)))........)))))))..))........ ( -29.50) >DroSim_CAF1 25102 111 + 1 GAUUAGCUGGCCCAAACUACAAUCGUGGGAGUAUUAAACUAUACGCUUGAUAG-GCUAAUUAAUAGAAGAAUUGAGCCGUUGGGUAACAUUACGGCUAGUCUUCGUAUUUAA (((((((((((((((....(((.((((..(((.....))).)))).)))...(-(((((((........)))).)))).)))))).......)))))))))........... ( -31.31) >DroYak_CAF1 25390 111 + 1 GUUUGGGUAGCCCAAACUACAAUCAUGGGAAUAUUUAACUCUACGCUUAAUGGAGGGAAUUAAUAGAAGAAUUAAACCAUUGGGUAAUAUUAUGGUUAGUUCUCGUAUGGA- (((((((...))))))).....(((((.((.((.((..(((((.......)))))..)).))......(((((((.((((...........))))))))))))).))))).- ( -26.00) >consensus GAUUAGCUGGCCCAAACUACAAUCGUGGGAGUAUUAAACUAUACGCUUGAUAG_GCUAAUUAAUAGAAGAAUUGAGCCGUUGGGUAACAUUAUGGCUAGUCUUCGUAUUUAA (((((((((((((((.((((....))))................(((((((....(((.....)))....)))))))..)))))).......)))))))))........... (-19.17 = -19.61 + 0.44)

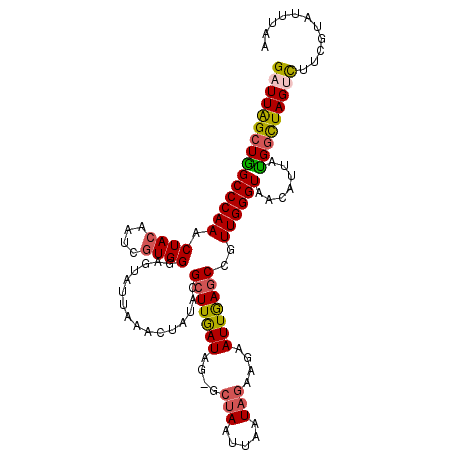
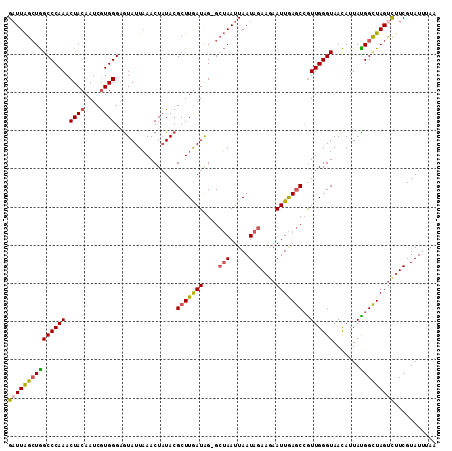
| Location | 3,064,379 – 3,064,490 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 87.44 |
| Mean single sequence MFE | -21.25 |
| Consensus MFE | -15.04 |
| Energy contribution | -15.85 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3064379 111 - 23771897 UUAAAUACGAAGACUAGCCAUAAUGUUACCCAACUGCUCAAUUCUUCUAUUAAUUAGC-CUAUCAAGCGUAUAGUUUAAUACUCCCACGAUUGUAGUUUGGGCCAGCUAAUC ...((((.(((((..(((......(((....))).)))....))))))))).((((((-...((((((.(((((((............)))))))))))))....)))))). ( -17.50) >DroSec_CAF1 25132 111 - 1 UUAAAUACGAAGGCUAGCCAUAAUGUUACCCAACGGCUCAAUUUUUCUAUUAAUUAGC-CUAUCAAGCGUAUAGUUUAAUACACCCACGAUUGUAGUUUGGGCCAGCUAAUC ...........((((.(((...............((((.((((........)))))))-)...(((((.(((((((............))))))))))))))).)))).... ( -22.90) >DroSim_CAF1 25102 111 - 1 UUAAAUACGAAGACUAGCCGUAAUGUUACCCAACGGCUCAAUUCUUCUAUUAAUUAGC-CUAUCAAGCGUAUAGUUUAAUACUCCCACGAUUGUAGUUUGGGCCAGCUAAUC ...((((.(((((..((((((...........))))))....))))))))).((((((-...((((((.(((((((............)))))))))))))....)))))). ( -25.50) >DroYak_CAF1 25390 111 - 1 -UCCAUACGAGAACUAACCAUAAUAUUACCCAAUGGUUUAAUUCUUCUAUUAAUUCCCUCCAUUAAGCGUAGAGUUAAAUAUUCCCAUGAUUGUAGUUUGGGCUACCCAAAC -....(((((......(((((...........)))))((((((((....(((((.......)))))....))))))))............)))))(((((((...))))))) ( -19.10) >consensus UUAAAUACGAAGACUAGCCAUAAUGUUACCCAACGGCUCAAUUCUUCUAUUAAUUAGC_CUAUCAAGCGUAUAGUUUAAUACUCCCACGAUUGUAGUUUGGGCCAGCUAAUC ........(((((..((((((...........))))))....)))))......(((((....((((((.(((((((............)))))))))))))....))))).. (-15.04 = -15.85 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:20 2006