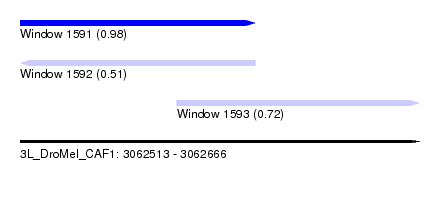
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,062,513 – 3,062,666 |
| Length | 153 |
| Max. P | 0.976119 |

| Location | 3,062,513 – 3,062,603 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 74.44 |
| Mean single sequence MFE | -21.85 |
| Consensus MFE | -10.79 |
| Energy contribution | -10.60 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
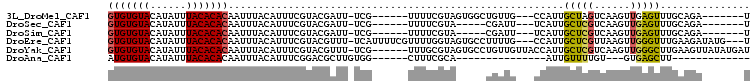
>3L_DroMel_CAF1 3062513 90 + 23771897 GUGUGUACAUAUUUACACACAAUUUACAUUUCGUACGAUU-UCG------UUUUCGUAGUGGCUGUUG---CCAUUGCUAGUCAAGUUGAGUUUGCAGA-------U (((((((......)))))))(((((((.....(.(((...-.))------)...)((((((((....)---)))))))..)).)))))...........-------. ( -23.20) >DroSec_CAF1 23257 85 + 1 GUGUGUACAUAUUUACACACAAUUUACAUUUCGUACGAUU-UCG------UUUUCGUA-----CGAUU---UCAUUGCUCGUCAAGUUGAGUUUGCAGA-------U (((((((......)))))))..........((((((((..-...------...)))))-----)))((---(((..(((((......))))).)).)))-------. ( -24.10) >DroSim_CAF1 22827 85 + 1 GUGUGUACAUAUUUACACACAAUUUACAUUUCGUACGAUU-UCG------UUUUCGUA-----CGAUU---UCAUUGCUCGUCAAGUUGAGUUUGCAGA-------U (((((((......)))))))..........((((((((..-...------...)))))-----)))((---(((..(((((......))))).)).)))-------. ( -24.10) >DroEre_CAF1 23971 100 + 1 GUGUGUACAUAUUUACACACAAUUUACAUUUCGUACGUUU-UCAUUUUCGUUUUGGUAGUGCCUUUUG---CCAUUGCUCGUUAAGUUGGGUUUGAAGAUAUG---U (((((((......)))))))................((((-(((..(((((..((((((......)))---)))..)).(.....)..)))..)))))))...---. ( -22.00) >DroYak_CAF1 23447 100 + 1 GUGUGUACAUAUUUACACACAAUUUACAUUUCGUACGUUU-UCG------UUUGCGUAGUGCCUGUUGUUACCAUUGCUCGUCAAGUUGGGCUUGAAGUUAUAUGAU (((((((......)))))))......((((.((((((...-.))------)..))).))))...........(((.(((..(((((.....))))))))...))).. ( -20.90) >DroAna_CAF1 23772 70 + 1 AUGUGUACAUAUUUACACACAAUUUACAUUUCGGACGCUUGUGG------CUUUCGCA---------------AUUGUUUUGU---GUGAGCUU------------- ...............(((((((...(((.((((((.((.....)------).)))).)---------------).))).))))---))).....------------- ( -16.80) >consensus GUGUGUACAUAUUUACACACAAUUUACAUUUCGUACGAUU_UCG______UUUUCGUA_____UGUUG___CCAUUGCUCGUCAAGUUGAGUUUGCAGA_______U (((((((......)))))))........................................................(((((......)))))............... (-10.79 = -10.60 + -0.19)
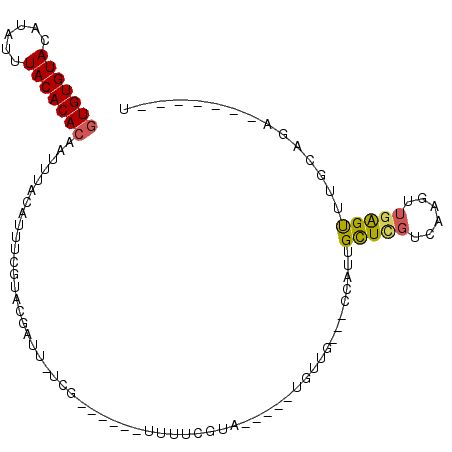
| Location | 3,062,513 – 3,062,603 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 74.44 |
| Mean single sequence MFE | -16.48 |
| Consensus MFE | -6.95 |
| Energy contribution | -6.82 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.42 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3062513 90 - 23771897 A-------UCUGCAAACUCAACUUGACUAGCAAUGG---CAACAGCCACUACGAAAA------CGA-AAUCGUACGAAAUGUAAAUUGUGUGUAAAUAUGUACACAC .-------..((((...((..............(((---(....)))).(((((...------...-..))))).))..))))....(((((((......))))))) ( -17.20) >DroSec_CAF1 23257 85 - 1 A-------UCUGCAAACUCAACUUGACGAGCAAUGA---AAUCG-----UACGAAAA------CGA-AAUCGUACGAAAUGUAAAUUGUGUGUAAAUAUGUACACAC .-------..((((..(((........)))......---..(((-----(((((...------...-..))))))))..))))....(((((((......))))))) ( -20.90) >DroSim_CAF1 22827 85 - 1 A-------UCUGCAAACUCAACUUGACGAGCAAUGA---AAUCG-----UACGAAAA------CGA-AAUCGUACGAAAUGUAAAUUGUGUGUAAAUAUGUACACAC .-------..((((..(((........)))......---..(((-----(((((...------...-..))))))))..))))....(((((((......))))))) ( -20.90) >DroEre_CAF1 23971 100 - 1 A---CAUAUCUUCAAACCCAACUUAACGAGCAAUGG---CAAAAGGCACUACCAAAACGAAAAUGA-AAACGUACGAAAUGUAAAUUGUGUGUAAAUAUGUACACAC (---(((.((...................((....)---)....((.....)).............-........)).)))).....(((((((......))))))) ( -12.00) >DroYak_CAF1 23447 100 - 1 AUCAUAUAACUUCAAGCCCAACUUGACGAGCAAUGGUAACAACAGGCACUACGCAAA------CGA-AAACGUACGAAAUGUAAAUUGUGUGUAAAUAUGUACACAC ....((((...(((((.....)))))((.((..((((..(....)..)))).))..(------((.-...))).))...))))....(((((((......))))))) ( -15.40) >DroAna_CAF1 23772 70 - 1 -------------AAGCUCAC---ACAAAACAAU---------------UGCGAAAG------CCACAAGCGUCCGAAAUGUAAAUUGUGUGUAAAUAUGUACACAU -------------.....(((---((((......---------------.((....)------).....((((.....))))...)))))))............... ( -12.50) >consensus A_______UCUGCAAACUCAACUUGACGAGCAAUGG___CAACA_____UACGAAAA______CGA_AAUCGUACGAAAUGUAAAUUGUGUGUAAAUAUGUACACAC .......................................................................................(((((((......))))))) ( -6.95 = -6.82 + -0.14)

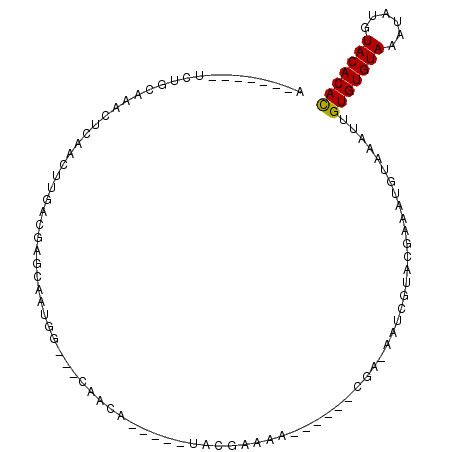
| Location | 3,062,573 – 3,062,666 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 83.72 |
| Mean single sequence MFE | -25.32 |
| Consensus MFE | -15.00 |
| Energy contribution | -15.44 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3062573 93 + 23771897 G---CCAUUGCUAGUCAAGUUGAGUUUGCAGA-------UACAUGGAUGUAUUUCCAGCUAUGUAGAUGUUAUCUAGCGCAUGAA--AACCCCCCGACUUGACCU .---.........(((((((((.(((((((..-------....((((......))))....)))))))(((.((........)).--)))....))))))))).. ( -23.60) >DroSec_CAF1 23312 95 + 1 U---UCAUUGCUCGUCAAGUUGAGUUUGCAGA-------UACAUGGAUGUAUUUCCAGCUUUGUAGAUAUUAUCUAGCGCCUGAAAAAACCCCCCGACUUGACCU .---.........(((((((((.(((((((((-------....((((......))))..)))))))))....((........))..........))))))))).. ( -23.40) >DroSim_CAF1 22882 95 + 1 U---UCAUUGCUCGUCAAGUUGAGUUUGCAGA-------UACAUGGAUGUAUUUCCAGCUUUGUAGAUAUUAUCUAGCGCCUGAAAAAACCCCCCGACUUGACCU .---.........(((((((((.(((((((((-------....((((......))))..)))))))))....((........))..........))))))))).. ( -23.40) >DroEre_CAF1 24037 94 + 1 G---CCAUUGCUCGUUAAGUUGGGUUUGAAGAUAUG---UAUAUGCAUGUAUUUCCAGCU----AGAUAUUAUCUAGCACCUGAA-AAACCCCCCGACUUGACCU .---.........(((((((((((...(((((((((---(....)))))).))))..(((----((((...))))))).......-......))))))))))).. ( -29.70) >DroYak_CAF1 23507 99 + 1 GUUACCAUUGCUCGUCAAGUUGGGCUUGAAGUUAUAUGAUAUAUGAAUGUAUUUCCAGCU----AGAUAUUAACUAGCGUCU-AA-AAACCCCCCGACUUGACCU .............(((((((((((...(((((.((((.........)))))))))..(((----((.......)))))....-..-......))))))))))).. ( -26.50) >consensus G___CCAUUGCUCGUCAAGUUGAGUUUGCAGA_______UACAUGGAUGUAUUUCCAGCU_UGUAGAUAUUAUCUAGCGCCUGAA_AAACCCCCCGACUUGACCU .............(((((((((.....................((((......))))(((....((((...)))))))................))))))))).. (-15.00 = -15.44 + 0.44)

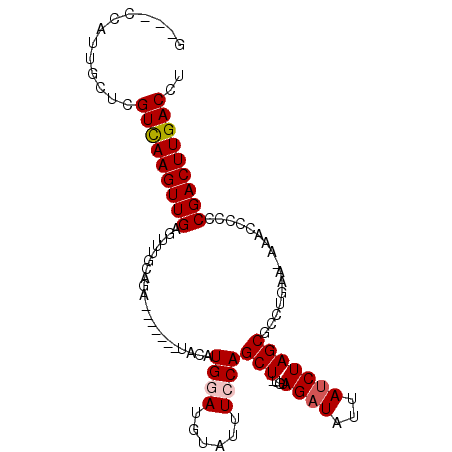
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:17 2006