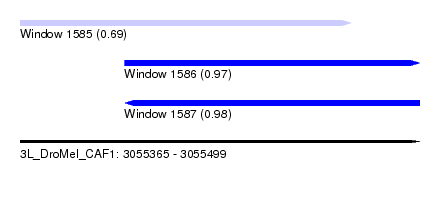
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,055,365 – 3,055,499 |
| Length | 134 |
| Max. P | 0.984041 |

| Location | 3,055,365 – 3,055,476 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.90 |
| Mean single sequence MFE | -30.73 |
| Consensus MFE | -22.36 |
| Energy contribution | -23.83 |
| Covariance contribution | 1.47 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3055365 111 + 23771897 --UUUUCGUUCCCUUCCCCUUUGCGUCCUCUUCCCCUUUUCCUAGCCAGCUGUUUGUCGCGUUCCACAAAUAUGGCUGGAGUGUGGAUCGCGUUUGGAGCCCAGGCUCCCACA --....................((((((.(........((((.(((((..(((((((........)))))))))))))))).).))).)))((..(((((....))))).)). ( -31.60) >DroSec_CAF1 16111 112 + 1 U-UUUUCGUUCCCUUCCCCUCUGCGUCCUCUUCCCCUCUUCCUAGCCAGCUGUUUGUCGCGUUCCACAAAUAUGGCUGGAGUGUGGAUCGCGCUUGUAGCCCAGGCUCCCACA .-....................(((......(((.(.((.((.(((((..(((((((........)))))))))))))))).).))).)))(((((.....)))))....... ( -26.50) >DroSim_CAF1 15865 113 + 1 UUUUUUCGUUCCCUUCCCCUCUGCGUCCUCUUCCCCUUUUCCUAGCCAGCUGUUUGUCGCGUUCCACAAAUAUGGCUGGAGUGUGGAUCGCGCUUGGAGCCCAGGCUCCCACA ......................((((.....(((.(..((((.(((((..(((((((........)))))))))))))))).).)))..))))..(((((....))))).... ( -32.30) >DroEre_CAF1 16243 113 + 1 CCUUUUCGUUCACUUCCCCGCUGCGCCCUCUUCCCCUCUUCCUAGCCAGCUGUUUGUCGCGUUCCACAAAUAUGGCUGGAGUGUGGACCGUGUUUGGAGCCCAGGCUCCCACA .......((((((((((((((((.((..................))))))(((((((........))))))).))..)))).)))))).(((...(((((....)))))))). ( -33.27) >DroYak_CAF1 16328 113 + 1 CCAUCUCAUUCCCUUCCCCGCUGCGCCCUCUUCCCCUUUUCCUAGCCAGCUGUUUGUCGCGUUCCACAAAUAUGGCUGGAGUGUGGAUCGCGCUUGGAGCCCAGGCUCCCACA ......................((((.....(((.(..((((.(((((..(((((((........)))))))))))))))).).)))..))))..(((((....))))).... ( -33.90) >DroPer_CAF1 28452 92 + 1 -----------UAUAUCCCCCUGCGCCGUAUCAAAAGUUUUGCAGCCAGCUGUUUUC--UGUUCCACAAAUAUGGCAGGAG--UGGAUCCCAUUUGG------GGGUCCGACA -----------........(((..(((((((....((....((((....))))...)--).........)))))))))).(--((((((((.....)------)))))).)). ( -26.82) >consensus __UUUUCGUUCCCUUCCCCUCUGCGCCCUCUUCCCCUUUUCCUAGCCAGCUGUUUGUCGCGUUCCACAAAUAUGGCUGGAGUGUGGAUCGCGCUUGGAGCCCAGGCUCCCACA ......................((((.....(((....((((.(((((..(((((((........))))))))))))))))...)))..))))..(((((....))))).... (-22.36 = -23.83 + 1.47)

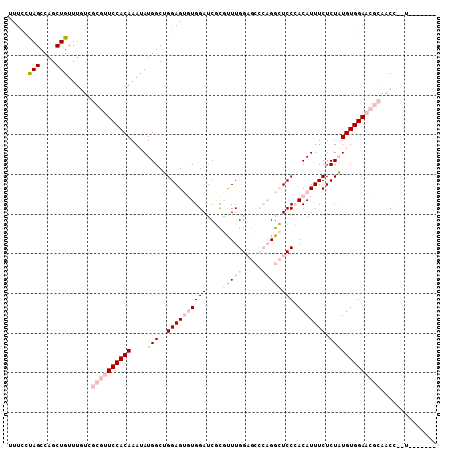
| Location | 3,055,400 – 3,055,499 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 79.19 |
| Mean single sequence MFE | -35.47 |
| Consensus MFE | -20.88 |
| Energy contribution | -23.77 |
| Covariance contribution | 2.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3055400 99 + 23771897 UUUCCUAGCCAGCUGUUUGUCGCGUUCCACAAAUAUGGCUGGAGUGUGGAUCGCGUUUGGAGCCCAGGCUCCCACAUUUCUCUAUGUGGAACGCAACC--U------- ..(((...((((((((((((........))))))..)))))).....)))..(((((((((((....)))))(((((......)))))))))))....--.------- ( -37.70) >DroPse_CAF1 24097 98 + 1 UUUUGCAGCCAGCUGUUUUC--UGUUCCACAAAUAUGGCAGGAG--UGGAUCCCAUUUGG------GGGUCCGACAUUUCUCUUUGUGGAUGUCGUCUGCUUUUUUGU ....((((....))))....--......(((((...((((((..--.(((((((.....)------))))))(((((((........))))))).))))))..))))) ( -32.70) >DroSim_CAF1 15902 99 + 1 UUUCCUAGCCAGCUGUUUGUCGCGUUCCACAAAUAUGGCUGGAGUGUGGAUCGCGCUUGGAGCCCAGGCUCCCACAUUUCUCUAUGUGGAACGCAACC--U------- .....(((....)))......((((((((((....(((..(((((((((...(.((((((...)))))).)))))))))).)))))))))))))....--.------- ( -37.20) >DroEre_CAF1 16280 99 + 1 CUUCCUAGCCAGCUGUUUGUCGCGUUCCACAAAUAUGGCUGGAGUGUGGACCGUGUUUGGAGCCCAGGCUCCCACAUUUCUCUAUGUGGAACGCAACC--U------- .....(((....)))......((((((((((....(((..(((((((((.(((....)))(((....))).))))))))).)))))))))))))....--.------- ( -36.00) >DroYak_CAF1 16365 99 + 1 UUUCCUAGCCAGCUGUUUGUCGCGUUCCACAAAUAUGGCUGGAGUGUGGAUCGCGCUUGGAGCCCAGGCUCCCACAUUUCUCUAUGUGGAACGCAACC--U------- .....(((....)))......((((((((((....(((..(((((((((...(.((((((...)))))).)))))))))).)))))))))))))....--.------- ( -37.20) >DroPer_CAF1 28478 97 + 1 UUUUGCAGCCAGCUGUUUUC--UGUUCCACAAAUAUGGCAGGAG--UGGAUCCCAUUUGG------GGGUCCGACAUUUCUCUUUGUGGAUGUUGUCUGCUUG-UUAU ....((((.((((.......--...(((((((....((.(((((--((((((((.....)------))))))...))))))))))))))).)))).))))...-.... ( -32.00) >consensus UUUCCUAGCCAGCUGUUUGUCGCGUUCCACAAAUAUGGCUGGAGUGUGGAUCGCGUUUGGAGCCCAGGCUCCCACAUUUCUCUAUGUGGAACGCAACC__U_______ .....(((....)))......((((((((((....(((..((((((((((....(((((.....)))))))).))))))).))))))))))))).............. (-20.88 = -23.77 + 2.89)

| Location | 3,055,400 – 3,055,499 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 79.19 |
| Mean single sequence MFE | -34.27 |
| Consensus MFE | -19.87 |
| Energy contribution | -23.53 |
| Covariance contribution | 3.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.984041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
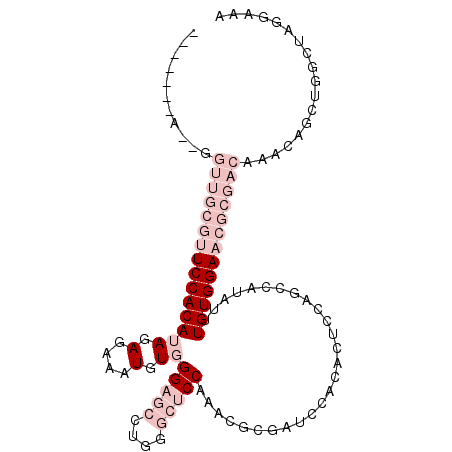
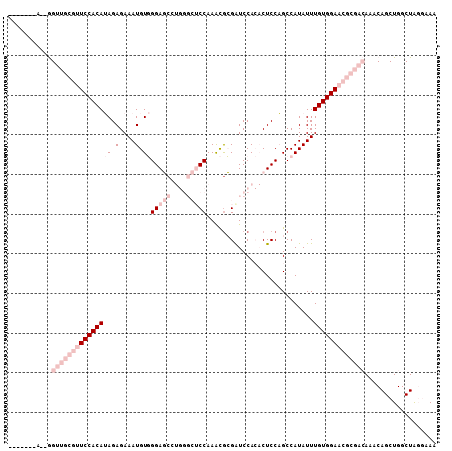
>3L_DroMel_CAF1 3055400 99 - 23771897 -------A--GGUUGCGUUCCACAUAGAGAAAUGUGGGAGCCUGGGCUCCAAACGCGAUCCACACUCCAGCCAUAUUUGUGGAACGCGACAAACAGCUGGCUAGGAAA -------.--(((((((((.(((((......)))))(((((....))))).))))))))).....((((((((..(((((.(....).)))))....))))).))).. ( -38.90) >DroPse_CAF1 24097 98 - 1 ACAAAAAAGCAGACGACAUCCACAAAGAGAAAUGUCGGACCC------CCAAAUGGGAUCCA--CUCCUGCCAUAUUUGUGGAACA--GAAAACAGCUGGCUGCAAAA ........((((.(....((((((((((......))(((.((------(.....))).))).--...........)))))))).((--(.......)))))))).... ( -25.60) >DroSim_CAF1 15902 99 - 1 -------A--GGUUGCGUUCCACAUAGAGAAAUGUGGGAGCCUGGGCUCCAAGCGCGAUCCACACUCCAGCCAUAUUUGUGGAACGCGACAAACAGCUGGCUAGGAAA -------.--(((((((((.(((((......)))))(((((....))))).))))))))).....((((((((..(((((.(....).)))))....))))).))).. ( -38.90) >DroEre_CAF1 16280 99 - 1 -------A--GGUUGCGUUCCACAUAGAGAAAUGUGGGAGCCUGGGCUCCAAACACGGUCCACACUCCAGCCAUAUUUGUGGAACGCGACAAACAGCUGGCUAGGAAG -------.--.(((((((((((((......((((((((((..((((((........))))))..)))...)))))))))))))))))))).................. ( -37.70) >DroYak_CAF1 16365 99 - 1 -------A--GGUUGCGUUCCACAUAGAGAAAUGUGGGAGCCUGGGCUCCAAGCGCGAUCCACACUCCAGCCAUAUUUGUGGAACGCGACAAACAGCUGGCUAGGAAA -------.--(((((((((.(((((......)))))(((((....))))).))))))))).....((((((((..(((((.(....).)))))....))))).))).. ( -38.90) >DroPer_CAF1 28478 97 - 1 AUAA-CAAGCAGACAACAUCCACAAAGAGAAAUGUCGGACCC------CCAAAUGGGAUCCA--CUCCUGCCAUAUUUGUGGAACA--GAAAACAGCUGGCUGCAAAA ....-...((((.(....((((((((((......))(((.((------(.....))).))).--...........)))))))).((--(.......)))))))).... ( -25.60) >consensus _______A__GGUUGCGUUCCACAUAGAGAAAUGUGGGAGCCUGGGCUCCAAACGCGAUCCACACUCCAGCCAUAUUUGUGGAACGCGACAAACAGCUGGCUAGGAAA ...........(((((((((((((((.(....).))(((((....)))))...........................))))))))))))).................. (-19.87 = -23.53 + 3.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:12 2006